4P9W
 
 | | Structure of ConA/Rh3Man | | Descriptor: | 2-[2-(2-{4-[(alpha-D-mannopyranosyloxy)methyl]-1H-1,2,3-triazol-1-yl}ethoxy)ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, CALCIUM ION, Concanavalin-A, ... | | Authors: | Sakai, F, Weiss, M.S, Jiang, M. | | Deposit date: | 2014-04-06 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of ConA/Rh3Man
To Be Published
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
4B4A
 
 | | Structure of the TatC core of the twin arginine protein translocation system | | Descriptor: | Lauryl Maltose Neopentyl Glycol, SEC-INDEPENDENT PROTEIN TRANSLOCASE PROTEIN TATC | | Authors: | Rollauer, S.E, Tarry, M.J, Jaaskelainen, M, Graham, J.E, Jaeger, F, Krehenbrink, M, Roversi, P, McDowell, M.A, Stansfeld, P.J, Johnson, S, Liu, S.M, Lukey, M.J, Marcoux, J, Robinson, C.V, Sansom, M.S, Palmer, T, Hogbom, M, Berks, B.C, Lea, S.M. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-05 | | Last modified: | 2012-12-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Tatc Core of the Twin-Arginine Protein Transport System.
Nature, 49, 2012
|
|
5FTU
 
 | | Tetrameric complex of Latrophilin 3, Unc5D and FLRT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.01 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
5FTT
 
 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
1C2N
 
 | | CYTOCHROME C2, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Cordier, F, Caffrey, M.S, Brutscher, B, Cusanovich, M.A, Marion, D, Blackledge, M. | | Deposit date: | 1998-04-27 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure, rotational diffusion anisotropy and local backbone dynamics of Rhodobacter capsulatus cytochrome c2.
J.Mol.Biol., 281, 1998
|
|
2EVN
 
 | | NMR solution structures of At1g77540 | | Descriptor: | Protein At1g77540 | | Authors: | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
6RTW
 
 | | Crystal structure of the Patched-1 (PTCH1) ectodomain in complex with nanobody NB64 and cholesterol-hemisuccinate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Llama-derived nanobody NB64, ... | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RVC
 
 | | Crystal structure of Patched-1 ectodomain 2 (PTCH1-ECD2) in complex with nanobody 75 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody NB75, Protein patched homolog 1, ... | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RVD
 
 | | Revised cryo-EM structure of the human 2:1 Ptch1-Shh complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | El Omari, K, Rudolf, A.F, Kowatsch, C, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RTX
 
 | | Crystal structure of the Patched-1 (PTCH1) ectodomain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RTY
 
 | | Crystal structure of the Patched ectodomain in complex with nanobody NB64 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Llama-derived nanobody NB64, Protein patched homolog 1 | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
3SNL
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 6-chloro-3,4-dimethyl-1-(3-methylpyridin-4-yl)-8-(trifluoromethyl)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
4V9B
 
 | | Crystal Structure of the 70S ribosome with tigecycline. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2012-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4VUB
 
 | | CCDB, A TOPOISOMERASE POISON FROM ESCHERICHIA COLI | | Descriptor: | CCDB, CHLORIDE ION | | Authors: | Loris, R, Dao-Thi, M.-H, Bahasi, E.M, Van Melderen, L, Poortmans, F, Liddington, R, Couturier, M, Wyns, L. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CcdB, a topoisomerase poison from E. coli.
J.Mol.Biol., 285, 1999
|
|
4W1R
 
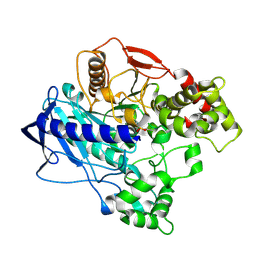 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 9.24 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-14 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4W1T
 
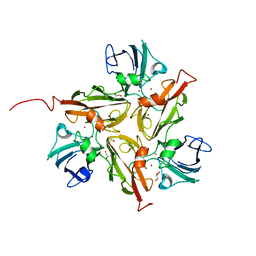 | |
4W8J
 
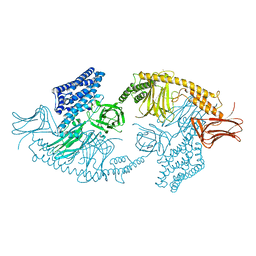 | | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pesticidal crystal protein cry1Ac | | Authors: | Evdokimov, A.G, Moshiri, F, Sturman, E.J, Rydel, T.J, Zheng, M, Seale, J.W, Franklin, S. | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals.
Protein Sci., 23, 2014
|
|
4V23
 
 | | RSV Matrix protein | | Descriptor: | MATRIX PROTEIN, POTASSIUM ION | | Authors: | Forster, A, Maertens, G, Bajorek, M. | | Deposit date: | 2014-10-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimerization of Matrix Protein is Required for Budding of Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4WBB
 
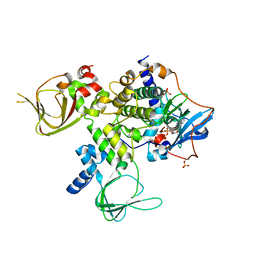 | | Single Turnover Autophosphorylation Cycle of the PKA RIIb Holoenzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Zhang, P, Knape, M.J, Ahuja, L.G, Keshwani, M.M, King, C.C, Sastri, M, Herberg, F.W, Taylor, S.S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single Turnover Autophosphorylation Cycle of the PKA RII beta Holoenzyme.
Plos Biol., 13, 2015
|
|
4V3H
 
 | | Crystal structure of CymA from Klebsiella oxytoca | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CYMA PROTEIN, NITRATE ION | | Authors: | van den Berg, B, Bhamidimarri, S.P, Kleinekathoefer, U, Winterhalter, M. | | Deposit date: | 2014-10-18 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Outer-membrane translocation of bulky small molecules by passive diffusion.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4UYZ
 
 | |
