6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6EHL
 
 | | Model of the Ebola virus nucleoprotein in recombinant nucleocapsid-like assemblies | | Descriptor: | Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
5OQO
 
 | | Crystal structure of the S. cerevisiae condensin Ycg1-Brn1 subcomplex bound to DNA (crystal form II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Condensin complex subunit 2, Condensin complex subunit 3, ... | | Authors: | Kschonsak, M, Hassler, M, Haering, C.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes.
Cell, 171, 2017
|
|
6ZVI
 
 | | Mbf1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
7A5J
 
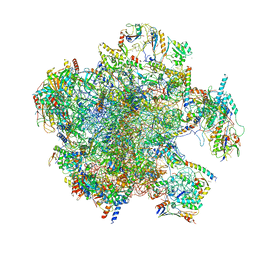 | | Structure of the split human mitoribosomal large subunit with P-and E-site mt-tRNAs | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
6ENS
 
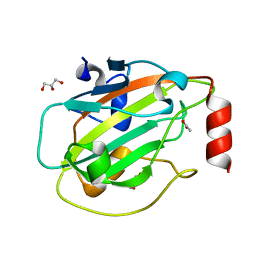 | | Structure of mouse wild-type RKIP | | Descriptor: | ACETATE ION, GLYCEROL, Phosphatidylethanolamine-binding protein 1 | | Authors: | Hirschbeck, M, Koelmel, W, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6EMX
 
 | |
7AAZ
 
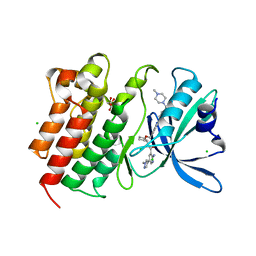 | | Crystal structure of MerTK in complex with a type 1.5 aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-~{N}-[(1~{S},2~{S})-2-[[4-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]methoxy]cyclopentyl]-5-(1-methylpyrazol-4-yl)pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
6EOG
 
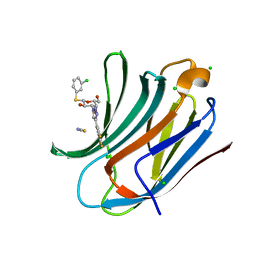 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-(3-chlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, Galectin-3, ... | | Authors: | Hakansson, M, Nilsson, U.J, Zetterberg, F, Logan, D.T. | | Deposit date: | 2017-10-09 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Monosaccharide Derivatives with Low-Nanomolar Lectin Affinity and High Selectivity Based on Combined Fluorine-Amide, Phenyl-Arginine, Sulfur-pi , and Halogen Bond Interactions.
ChemMedChem, 13, 2018
|
|
6EOU
 
 | |
6ZN6
 
 | | Protein polybromo-1 (PB1 BD2) Bound To MW278 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-azanyl-5-[(3~{R})-3-phenoxypiperidin-1-yl]pyridazin-3-yl]phenol, Protein polybromo-1 | | Authors: | Preuss, F, Mathea, S, Chatterjee, D, Wanior, M, Joerger, A.C, Knapp, S. | | Deposit date: | 2020-07-06 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
4RMG
 
 | | Human Sirt2 in complex with SirReal2 and NAD+ | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
6ZOK
 
 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6E5W
 
 | | Crystal structure of human cellular retinol binding protein 3 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, GLYCEROL, Retinol-binding protein 5 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6E6Q
 
 | | 1.20 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | Deposit date: | 2018-07-25 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
5OPY
 
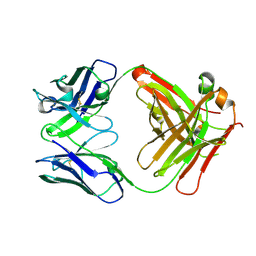 | | Crystal structure of anti-alphaVbeta3 integrin Fab LM609 | | Descriptor: | Heavy chain of LM609 Fab (antigen-binding fragment), Light chain of LM609 Fab (antigen-binding fragment) | | Authors: | Backovic, M, Veesler, D, Borst, A.J, James, Z.M, Zagotta, W, Ginsberg, M, Rey, F.A, DiMaio, F. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
6ELV
 
 | | Recombinantly expressed C-terminal domain of MdPPO1 (Csole-domain) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Polyphenol oxidase, ... | | Authors: | Kampatsikas, I, Bijelic, A, Pretzler, M, Rompel, A. | | Deposit date: | 2017-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A Peptide-Induced Self-Cleavage Reaction Initiates the Activation of Tyrosinase.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6E6R
 
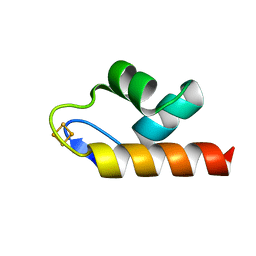 | | 1.50 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) R26E mutant from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | Deposit date: | 2018-07-25 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
6EO5
 
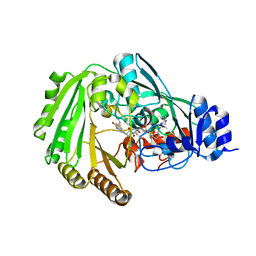 | | Physcomitrella patens BBE-like 1 variant D396N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PpBBE-like 1 D396N | | Authors: | Toplak, M, Winkler, A, Macheroux, P. | | Deposit date: | 2017-10-09 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The single berberine bridge enzyme homolog of Physcomitrella patens is a cellobiose oxidase.
FEBS J., 285, 2018
|
|
6EOI
 
 | | Reductive Aminase from Aspergillus terreus in complex with NADPH and ethyl-5-oxohexanoate | | Descriptor: | MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reductive Aminase, ... | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Mechanism for Reductive Amination Catalyzed by Fungal Reductive Aminases
Acs Catalysis, 2018
|
|
5OQN
 
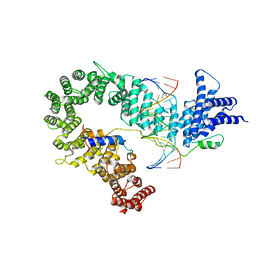 | |
6EGX
 
 | | Sacbrood virus of honeybee - expansion state I | | Descriptor: | minor capsid protein MiCP, structural protein VP1, structural protein VP2, ... | | Authors: | Plevka, P, Prochazkova, M. | | Deposit date: | 2017-09-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Virion structure and genome delivery mechanism of sacbrood honeybee virus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E6K
 
 | | Crystal structure of human cellular retinol-binding protein 4 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinoid-binding protein 7 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
5OSH
 
 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL N-terminal domain (1-236) | | Descriptor: | Interaptin, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8DVB
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (1'-(4-((1-butylpyrrolidin-3-yl)methoxy)phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-1'-(4-{[(3R)-1-(3-fluoropropyl)pyrrolidin-3-yl]methoxy}phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
