4AEI
 
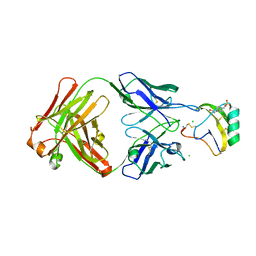 | | Crystal structure of the AaHII-Fab4C1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALPHA-MAMMAL TOXIN AAH2, CHLORIDE ION, ... | | Authors: | Fabrichny, I.P, Mondielli, G, Conrod, S, Martin-Eauclaire, M.F, Bourne, Y, Marchot, P. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into Antibody Sequestering and Neutralizing of Na+-Channel & [Alpha]-Type Modulator from Old-World Scorpion Venom
J.Biol.Chem., 287, 2012
|
|
4B5Q
 
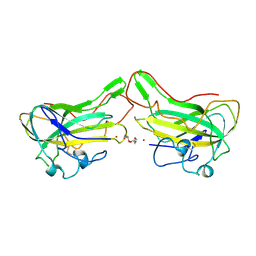 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | Descriptor: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | Authors: | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
2MAR
 
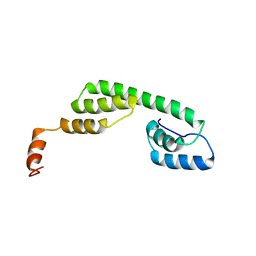 | | Solution structure of Ani s 5 Anisakis simplex allergen | | Descriptor: | SXP/RAL-2 family protein | | Authors: | Garcia-Mayoral, M.F, Trevino, M.A, Perez-Pinar, T, Caballero, M.L, Knaute, T, Umpierrez, A, Bruix, M, Rodriguez-Perez, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Relationships between IgE/IgG4 epitopes, structure and function in Anisakis simplex Ani s 5, a member of the SXP/RAL-2 protein family
Plos Negl Trop Dis, 8, 2014
|
|
4AY6
 
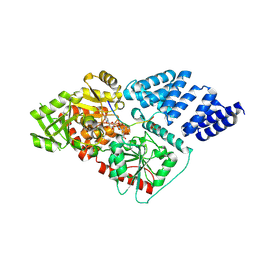 | | Human O-GlcNAc transferase (OGT) in complex with UDP-5SGlcNAc and substrate peptide | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, SULFATE ION, TGF-BETA-ACTIVATED KINASE 1 AND MAP3K7-BINDING PROTEIN 1, ... | | Authors: | Schimpl, M, Zheng, X, Blair, D.E, Schuettelkopf, A.W, Navratilova, I, Aristotelous, T, Ferenbach, A.T, Macnaughtan, M.A, Borodkin, V.S, van Aalten, D.M.F. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-24 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | O-Glcnac Transferase Invokes Nucleotide Sugar Pyrophosphate Participation in Catalysis
Nat.Chem.Biol., 8, 2012
|
|
4CXS
 
 | | G4 mutant of PAS, arylsulfatase from Pseudomonas aeruginosa, in complex with Phenylphosphonic acid | | Descriptor: | ARYLSULFATASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-08 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2K4I
 
 | | Solution structure of HIV-2 myrMA bound to di-C4-PI(4,5)P2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1,2-DIYL DIBUTANOATE, HIV-2 myristoylated matrix protein, MYRISTIC ACID | | Authors: | Saad, J.S, Ablan, S.D, Ghanam, R.H, Kim, A, Andrews, K, Nagashima, K, Freed, E.O, Summers, M.F. | | Deposit date: | 2008-06-08 | | Release date: | 2008-08-12 | | Last modified: | 2014-01-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting.
J.Mol.Biol., 382, 2008
|
|
4BHU
 
 | | Crystal structure of BslA - A bacterial hydrophobin | | Descriptor: | CHLORIDE ION, GLYCEROL, UNCHARACTERIZED PROTEIN YUAB | | Authors: | Rao, F.V, Hobley, L, Ostrowski, A, Bromley, K.M, Porter, M, Prescott, A.R, Swedlow, J.R, MacPhee, C.E, van Aalten, D.M.F, Stanley-Wall, N.R. | | Deposit date: | 2013-04-08 | | Release date: | 2013-08-14 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Bsla is a Self-Assembling Bacterial Hydrophobin that Coats the Bacillus Subtilis Biofilm.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2JPR
 
 | | Joint refinement of the HIV-1 CA-NTD in complex with the assembly inhibitor CAP-1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, Gag-Pol polyprotein | | Authors: | Kelly, B.N, Kyere, S, Kinde, I, Tang, C, Howard, B.R, Robinson, H, Sundquist, W.I, Summers, M.F, Hill, C.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein
J.Mol.Biol., 373, 2007
|
|
2JZ3
 
 | | SOCS box elonginBC ternary complex | | Descriptor: | Suppressor of cytokine signaling 3, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2 | | Authors: | Babon, J.J, Sabo, J, Soetopo, A, Yao, S, Bailey, M.F, Zhang, J, Nicola, N.A, Norton, R.S. | | Deposit date: | 2007-12-27 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase
J.Mol.Biol., 381, 2008
|
|
4CYR
 
 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4BSM
 
 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.5A | | Descriptor: | EXPORTIN-1 | | Authors: | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
4BQH
 
 | | Crystal structure of the uridine diphosphate N-acetylglucosamine pyrophosphorylase from Trypanosoma brucei in complex with inhibitor | | Descriptor: | (3S)-3-[2-(1,3-benzodioxol-5-yl)-2-oxidanylidene-ethyl]-4-bromanyl-5-methyl-3-oxidanyl-1H-indol-2-one, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Fang, W, Raimi, O.G, vanAalten, D.M.F. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Allosteric Inhibitor of the Uridine Diphosphate N-Acetylglucosamine Pyrophosphorylase from Trypanosoma Brucei.
Acs Chem.Biol., 8, 2013
|
|
2J9X
 
 | | Tryptophan Synthase in complex with GP, alpha-D,L-glycerol-phosphate, Cs, pH6.5 - alpha aminoacrylate form - (GP)E(A-A) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ngo, H, Kimmich, N, Harris, R, Niks, D, Blumenstein, L, Kulik, V, Barends, T.R, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-11-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Regulation of Substrate Channeling in Tryptophan Synthase: Modulation of the L-Serine Reaction in Stage I of the Beta-Reaction by Alpha-Site Ligands.
Biochemistry, 46, 2007
|
|
2K4E
 
 | | Solution structure of the HIV-2 UNMYRISTOYLATED MATRIX PROTEIN | | Descriptor: | HIV-2 unmyristoylated matrix protein | | Authors: | Saad, J.S, Ablan, S.D, Ghanam, R.H, Kim, A, Andrews, K, Nagashima, K, Freed, E.O, Summers, M.F. | | Deposit date: | 2008-06-07 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting.
J.Mol.Biol., 382, 2008
|
|
2JLB
 
 | | Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, URIDINE-DIPHOSPHATE-METHYLENE-N-ACETYL-GLUCOSAMINE, ... | | Authors: | Schuettelkopf, A.W, Clarke, A.J, van Aalten, D.M.F. | | Deposit date: | 2008-09-07 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into Mechanism and Specificity of O-Glcnac Transferase.
Embo J., 27, 2008
|
|
2JVZ
 
 | | Solution NMR Structure of the Second and Third KH Domains of KSRP | | Descriptor: | Far upstream element-binding protein 2 | | Authors: | Diaz-Moreno, I, Hollingworth, D, Garcia-Mayoral, M.F, Kelly, G, Cukier, C.D, Ramos, A. | | Deposit date: | 2007-09-28 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Second and Third KH Domains of KSRP
To be Published, 2007
|
|
2L1F
 
 | | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography | | Descriptor: | RNA (65-MER), RNA (66-MER) | | Authors: | Summers, M.F, Irobalieva, R.N, Tolbert, B, Smalls-Manty, A, Iyalla, K, Loeliger, K, D'Souza, V, Khant, H, Schmid, M, Garcia, E, Telesnitsky, A, Chiu, W, Miyazaki, Y. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography.
J.Mol.Biol., 404, 2010
|
|
4AY1
 
 | | Human YKL-39 is a pseudo-chitinase with retained chitooligosaccharide binding properties | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE-3-LIKE PROTEIN 2 | | Authors: | Schimpl, M, Rush, C.L, Betou, M, Eggleston, I.M, Penman, G.A, Recklies, A.D, van Aalten, D.M.F. | | Deposit date: | 2012-06-17 | | Release date: | 2012-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human Ykl-39 is a Pseudo-Chitinase with Retained Chitooligosaccharide-Binding Properties.
Biochem.J., 446, 2012
|
|
4AY5
 
 | | Human O-GlcNAc transferase (OGT) in complex with UDP and glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GTAB1TIDE, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYL TRANSFERASE 110 KDA SUBUNIT, ... | | Authors: | Schimpl, M, Zheng, X, Blair, D.E, Schuettelkopf, A.W, Navratilova, I, Aristotelous, T, Ferenbach, A.T, Macnaughtan, M.A, Borodkin, V.S, van Aalten, D.M.F. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | O-Glcnac Transferase Invokes Nucleotide Sugar Pyrophosphate Participation in Catalysis
Nat.Chem.Biol., 8, 2012
|
|
4CXU
 
 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with 3-Br-Phenolphenylphosphonate | | Descriptor: | 3-bromophenyl hydrogen (S)-phenylphosphonate, ARYLSULFATASE, CALCIUM ION | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CYS
 
 | | G6 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with Phenylphosphonic acid | | Descriptor: | AMMONIUM ION, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2N1Q
 
 | | HIV-1 Core Packaging Signal | | Descriptor: | RNA_(155-MER) | | Authors: | Keane, S.C, Summers, M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA structure. Structure of the HIV-1 RNA packaging signal.
Science, 348, 2015
|
|
2N0M
 
 | | The solution structure of the soluble form of the Lipid-modified Azurin from Neisseria gonorrhoeae | | Descriptor: | COPPER (I) ION, Lipid modified azurin protein | | Authors: | Pauleta, S.R, Matzapetakis, M.F, Nobrega, C.F, Carreira, C, Saraiva, I.H. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the soluble form of the lipid-modified azurin from Neisseria gonorrhoeae, the electron donor of cytochrome c peroxidase.
Biochim.Biophys.Acta, 1857, 2016
|
|
4A0O
 
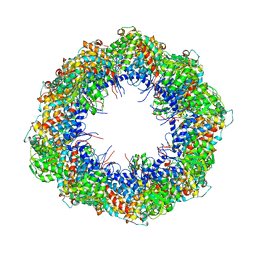 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A82
 
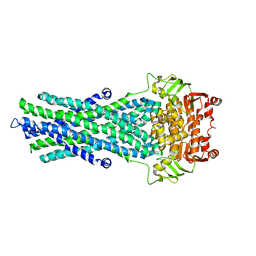 | | Fitted model of staphylococcus aureus sav1866 model ABC transporter in the human cystic fibrosis transmembrane conductance regulator volume map EMD-1966. | | Descriptor: | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR | | Authors: | Rosenberg, M.F, ORyan, L.P, Hughes, G, Zhao, Z, Aleksandrov, L.A, Riordan, J.R, Ford, R.C. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | The Cystic Fibrosis Transmembrane Conductance Regulator (Cftr):3D Structure and Localisation of a Channel Gate.
J.Biol.Chem., 286, 2011
|
|
