5MEI
 
 | | Crystal structure of Agelastatin A bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | McClary, B, Zinshteyn, B, Meyer, M, Jouanneau, M, Pellegrino, S, Yusupova, G, Schuller, A, Reyes, J.C.P, Lu, J, Luo, C, Dang, Y, Romo, D, Yusupov, M, Green, R, Liu, J.O. | | Deposit date: | 2016-11-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of Eukaryotic Translation by the Antitumor Natural Product Agelastatin A.
Cell Chem Biol, 24, 2017
|
|
4GXO
 
 | | Crystal structure of Staphylococcus aureus protein SarZ mutant C13E | | Descriptor: | GLYCEROL, MarR family regulatory protein | | Authors: | Sun, F, Ding, Y, Ji, Q, Liang, Z, Deng, X, Wong, C.C, Yi, C, Zhang, L, Xie, S, Alvarez, S, Hicks, L.M, Luo, C, Jiang, H, Lan, L, He, C. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6AFR
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with 5-((4-fluoro-1H-imidazol-1-yl)methyl)quinolin-8-ol | | Descriptor: | 5-[(4-fluoranylimidazol-1-yl)methyl]quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Xing, J, Zhang, R.K, Zheng, M.Y, Luo, C, Jiang, X.R. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Rational design of 5-((1H-imidazol-1-yl)methyl)quinolin-8-ol derivatives as novel bromodomain-containing protein 4 inhibitors
Eur J Med Chem, 163, 2018
|
|
7CPD
 
 | | Crystal structure of T2R-TTL-(+)-6-Br-JP18 complex | | Descriptor: | (6R)-6-[(6-bromanyl-1H-indol-3-yl)methyl]-6,7,8,9-tetrahydrobenzo[7]annulen-5-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2020-08-06 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal structure of T2R-TTL-(+)-6-Br-JP18 complex
To Be Published
|
|
7CPQ
 
 | | crystal structure of T2R-TTL-(+)-6-Cl-JP18 complex | | Descriptor: | (6R)-6-[(6-chloranyl-1H-indol-3-yl)methyl]-6,7,8,9-tetrahydrobenzo[7]annulen-5-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2020-08-07 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | crystal structure of T2R-TTL-(+)-6-Cl-JP18 complex
To Be Published
|
|
4EMP
 
 | | Crystal structure of the mutant of ClpP E137A from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Ye, F, Zhang, J, Liu, H, Luo, C, Yang, C.-G. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
J.Biol.Chem., 288, 2013
|
|
6CV0
 
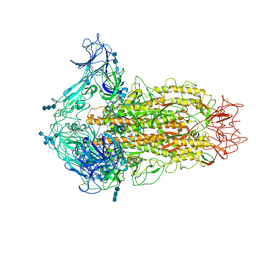 | | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Luo, C, Zhang, W, Li, F. | | Deposit date: | 2018-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins.
PLoS Pathog., 14, 2018
|
|
1IA8
 
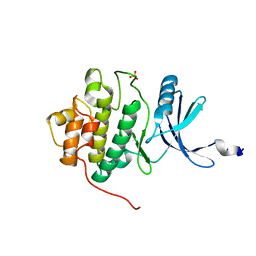 | | THE 1.7 A CRYSTAL STRUCTURE OF HUMAN CELL CYCLE CHECKPOINT KINASE CHK1 | | Descriptor: | CHK1 CHECKPOINT KINASE, SULFATE ION | | Authors: | Chen, P, Luo, C, Deng, Y, Ryan, K, Register, J, Margosiak, S, Tempczyk-Russell, A, Nguyen, B, Myers, P, Lundgren, K, Chen Kan, C.-C, O'Connor, P.M. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation.
Cell(Cambridge,Mass.), 100, 2000
|
|
9B7A
 
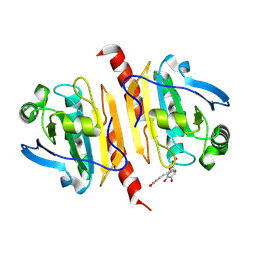 | | Crystal structure of peroxiredoxin 1 with RA | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, Peroxiredoxin-1 | | Authors: | Wu, Y, Xu, H, Luo, C. | | Deposit date: | 2024-03-27 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inhibited peroxidase activity of peroxiredoxin 1 by palmitic acid exacerbates nonalcoholic steatohepatitis in male mice.
Nat Commun, 16, 2025
|
|
7WZC
 
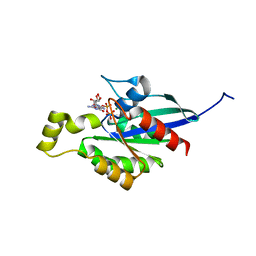 | | An open conformation Form2 of switch II for RhoA GDP-bound state | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79944921 Å) | | Cite: | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
7WZA
 
 | | An open conformation Form 1 of switch II for RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50028777 Å) | | Cite: | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
5Y1Y
 
 | |
6KX2
 
 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
5H21
 
 | | Trimethoxy-ring inhibitor in complex with the first bromodomain of BRD4 | | Descriptor: | 3,4,5-trimethoxy-~{N}-(2-thiophen-2-ylethyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of novel trimethoxy-ring BRD4 bromodomain inhibitors: AlphaScreen assay, crystallography and cell-based assay.
Medchemcomm, 8, 2017
|
|
7WEU
 
 | |
7WET
 
 | | Crystal structure of Peroxiredoxin I in complex with the inhibitor Cela | | Descriptor: | (2R,4aS,6aS,12bR,14aS,14bR)-10-hydroxy-2,4a,6a,9,12b,14a-hexamethyl-11-oxo-1,2,3,4,4a,5,6,6a,11,12b,13,14,14a,14b-tetradecahydropicene-2-carboxylic acid, Peroxiredoxin-1 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2021-12-24 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Celastrol suppresses colorectal cancer via covalent targeting peroxiredoxin 1.
Signal Transduct Target Ther, 8, 2023
|
|
5XAF
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
6LQX
 
 | | Crystal structure of the CBP bromodomain in complex with small molecule LC-CPin7 | | Descriptor: | (1~{S},6~{R})-6-[(1-methoxycarbonyl-3,4-dihydro-2~{H}-quinolin-6-yl)carbamoyl]cyclohex-3-ene-1-carboxylic acid, CREB-binding protein, SODIUM ION | | Authors: | Chen, Y, Zhang, F, Sun, Z, Bi, X, Luo, C. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Design, synthesis and biological evaluation of novel small molecule inhibitor of the CBP bromodomain with possible anti-leukemia effects
To Be Published
|
|
6LU5
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-25 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86527729 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6LU6
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5-2 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-26 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.970063 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
5Z5T
 
 | | The first bromodomain of BRD4 with compound BDF-2141 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z5V
 
 | | The first bromodomain of BRD4 with compound BDF-1253 | | Descriptor: | Bromodomain-containing protein 4, N-{8-hydroxy-4-[(1H-imidazol-1-yl)methyl]quinolin-2-yl}acetamide | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z5U
 
 | | The first bromodomain of BRD4 with compound BDF-2254 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinoline-6,8-diol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z4P
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound TCA complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6,7,8-trimethoxy-1-(4-methoxyphenyl)-4,5-dihydro-2~{H}-benzo[e]indazole, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-12 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Design of novel Tubulin inhibitors
To Be Published
|
|
