7EAO
 
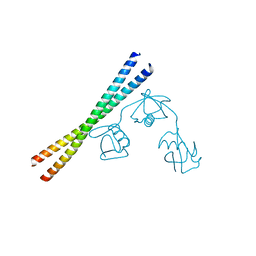 | |
5H07
 
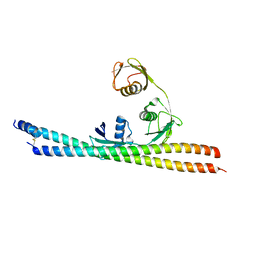 | | TNIP2-Ub complex, C2 form | | Descriptor: | Polyubiquitin-C, TNFAIP3-interacting protein 2 | | Authors: | Lo, Y.C, Lin, S.C. | | Deposit date: | 2016-10-04 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Structural Insights into Linear Tri-ubiquitin Recognition by A20-Binding Inhibitor of NF-kappa B, ABIN-2
Structure, 25, 2017
|
|
2VYE
 
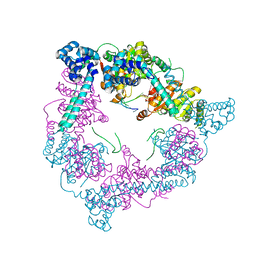 | | Crystal Structure of the DnaC-ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP)-3', REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
4APV
 
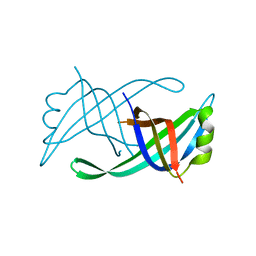 | | The Klebsiella pneumoniae primosomal PriB protein: identification, crystal structure, and ssDNA binding mode | | Descriptor: | PRIMOSOMAL REPLICATION PROTEIN N | | Authors: | Lo, Y.H, Huang, Y.H, Hsiao, C.D, Huang, C.Y. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal Structure and DNA-Binding Mode of Klebsiella Pneumoniae Primosomal Prib Protein.
Genes Cells, 17, 2012
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7T0V
 
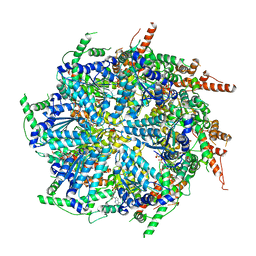 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
3MOP
 
 | | The ternary Death Domain complex of MyD88, IRAK4, and IRAK2 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, Interleukin-1 receptor-associated kinase-like 2, Myeloid differentiation primary response protein MyD88 | | Authors: | Lin, S.-C, Lo, Y.-C, Wu, H. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling.
Nature, 465, 2010
|
|
8DWT
 
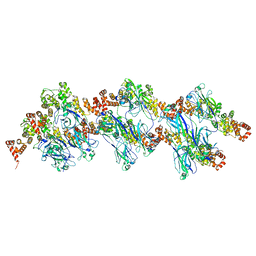 | | SPOP W22R Form 2 | | Descriptor: | Speckle-type POZ protein | | Authors: | Cuneo, M.J, Mittag, T, O'Flynn, B, Lo, Y.H. | | Deposit date: | 2022-08-02 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation.
Mol.Cell, 83, 2023
|
|
8DWV
 
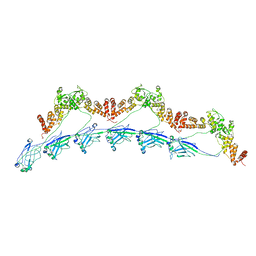 | | Full-length wild type SPOP | | Descriptor: | Speckle-type POZ protein | | Authors: | Cuneo, M.J, Mittag, T, O'Flynn, B, Lo, Y.H. | | Deposit date: | 2022-08-02 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation.
Mol.Cell, 83, 2023
|
|
8DWS
 
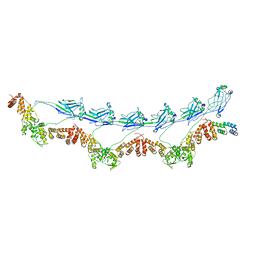 | | Full-length E47K SPOP | | Descriptor: | Speckle-type POZ protein | | Authors: | Cuneo, M.J, Mittag, T, O'Flynn, B, Lo, Y.H. | | Deposit date: | 2022-08-02 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation.
Mol.Cell, 83, 2023
|
|
3HCT
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the P1 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCU
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the C2 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCS
 
 | | Crystal structure of the N-terminal domain of TRAF6 | | Descriptor: | TNF receptor-associated factor 6, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3QA8
 
 | | Crystal Structure of inhibitor of kappa B kinase beta | | Descriptor: | MGC80376 protein | | Authors: | Xu, G, Lo, Y.C, Li, Q, Napolitano, G, Wu, X, Jiang, X, Dreano, M, Karin, M, Wu, H. | | Deposit date: | 2011-01-10 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of inhibitor of kappa B kinase beta.
Nature, 472, 2011
|
|
3RZF
 
 | | Crystal Structure of Inhibitor of kappaB kinase beta (I4122) | | Descriptor: | (4-{[4-(4-chlorophenyl)pyrimidin-2-yl]amino}phenyl)[4-(2-hydroxyethyl)piperazin-1-yl]methanone, MGC80376 protein | | Authors: | Xu, G, Lo, Y.C, Li, Q, Napolitano, G, Wu, X, Jiang, X, Dreano, M, Karin, M, Wu, H. | | Deposit date: | 2011-05-11 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor of KappaB kinase Beta.
Nature, 472, 2011
|
|
3DKB
 
 | | Crystal Structure of A20, 2.5 angstrom | | Descriptor: | Tumor necrosis factor, alpha-induced protein 3 | | Authors: | Lin, S.-C, Chung, J.Y, Lo, Y.-C, Wu, H. | | Deposit date: | 2008-06-24 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20
J.Mol.Biol., 376, 2008
|
|
2W58
 
 | | Crystal Structure of the DnaI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, K.L, Lo, Y.H, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Interplay between the Replicative Helicase Dnac and its Loader Protein Dnai from Geobacillus Kaustophilus.
J.Mol.Biol., 393, 2009
|
|
6TXR
 
 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5WVC
 
 | | Structure of the CARD-CARD disk | | Descriptor: | Apoptotic protease-activating factor 1, Caspase, IODIDE ION | | Authors: | Lin, S.C, Lo, Y.C, Su, T.W. | | Deposit date: | 2016-12-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Structural Insights into DD-Fold Assembly and Caspase-9 Activation by the Apaf-1 Apoptosome.
Structure, 25, 2017
|
|
1VF2
 
 | |
1VF3
 
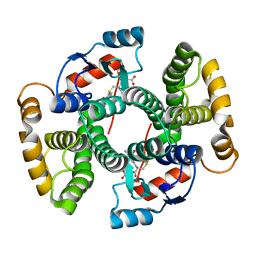 | | cGSTA1-1 in complex with glutathione conjugate of CDNB | | Descriptor: | ACETIC ACID, GLUTATHIONE S-(2,4 DINITROBENZENE), Glutathione S-transferase 3 | | Authors: | Lin, S.C, Lo, Y.C, Tam, M.F, Liaw, Y.C. | | Deposit date: | 2004-04-07 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of chicken glutathione S-transferase A1-1
To be Published
|
|
3PB4
 
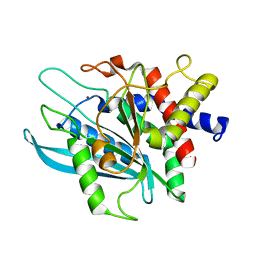 | | Crystal structure of the catalytic domain of human Golgi-resident glutaminyl cyclase at pH 6.0 | | Descriptor: | Glutaminyl-peptide cyclotransferase-like protein, ZINC ION | | Authors: | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
3PBB
 
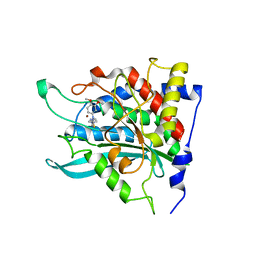 | | Crystal structure of human secretory glutaminyl cyclase in complex with PBD150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
3PB6
 
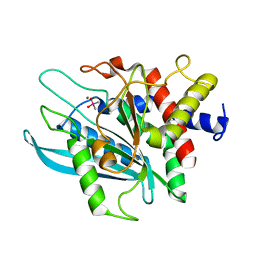 | | Crystal structure of the catalytic domain of human Golgi-resident glutaminyl cyclase at pH 6.5 | | Descriptor: | CACODYLATE ION, Glutaminyl-peptide cyclotransferase-like protein, ZINC ION | | Authors: | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
3PB7
 
 | | Crystal structure of the catalytic domain of human Golgi-resident glutaminyl cyclase in complex with PBD150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl-peptide cyclotransferase-like protein, ZINC ION | | Authors: | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
