7YOS
 
 | |
1SN1
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
3LEH
 
 | |
5F5M
 
 | | Crystal structure of Marburg virus nucleoprotein core domain | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | Authors: | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1CS7
 
 | | SYNTHETIC DNA HAIRPIN WITH STILBENEDIETHER LINKER | | Descriptor: | 5'-D(GP*(BRU)P*TP*TP*TP*GP*(S02)*CP*AP*AP*AP*AP*C)-3', STRONTIUM ION | | Authors: | Lewis, F.D, Liu, X, Wu, Y, Miller, S.E, Wasielewski, M.R, Letsinger, R.L, Sanishvili, R, Joachimiak, A, Tereshko, V, Egli, M. | | Deposit date: | 1999-08-17 | | Release date: | 2001-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Photoinduced Electron Transfer in Exceptionally Stable Synthetic DNA Hairpins with Stilbenediether Linkers
J.Am.Chem.Soc., 121, 1999
|
|
5GT2
 
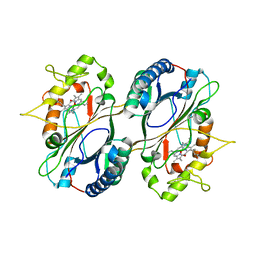 | | Crystal Structure and Biochemical Features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable deferrochelatase/peroxidase YfeX | | Authors: | Ma, Y.L, Yuan, Z.G, Liu, S, Wang, J.X, Gu, L.C, Liu, X.H. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates
Biochem. Biophys. Res. Commun., 484, 2017
|
|
8WEJ
 
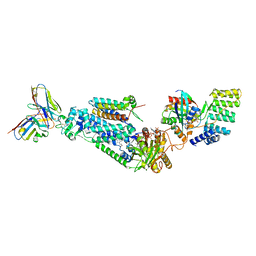 | |
8X2L
 
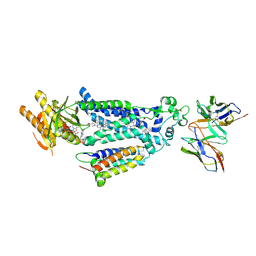 | |
5JPP
 
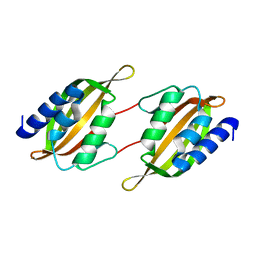 | | Structure of limonene epoxide hydrolase mutant - H-2-H5 | | Descriptor: | limonene epoxide hydrolase | | Authors: | Li, G, Zhang, H, Sun, Z, Liu, X, Reetz, M.T. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-Parameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity and Activity of an Epoxide Hydrolase
To be published
|
|
5KJH
 
 | |
5KKL
 
 | | Structure of ctPRC2 in complex with H3K27me3 and H3K27M | | Descriptor: | ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA, Putative uncharacterized protein,Histone H3.1 peptide,Zinc finger domain-containing protein, S-ADENOSYLMETHIONINE, ... | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2016-06-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Response to Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Science, 354, 2016
|
|
5KJI
 
 | | Crystal structure of an active polycomb repressive complex 2 in the basal state | | Descriptor: | Putative uncharacterized protein,Zinc finger domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2016-06-20 | | Release date: | 2017-04-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Response to Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Science, 354, 2016
|
|
7BU7
 
 | | Structure of human beta1 adrenergic receptor bound to BI-167107 and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-04 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
7BVQ
 
 | | Structure of human beta1 adrenergic receptor bound to carazolol | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
7BU6
 
 | | Structure of human beta1 adrenergic receptor bound to norepinephrine and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl octanoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-04 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
7BTS
 
 | | Structure of human beta1 adrenergic receptor bound to epinephrine and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
5F5O
 
 | | Crystal structure of Marburg virus nucleoprotein core domain bound to VP35 regulation peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35, SULFATE ION | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
3WFV
 
 | | HIV-1 CRF07 gp41 | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Du, J, Xue, H, Ma, J, Liu, F, Zhou, J, Shao, Y, Qiao, W, Liu, X. | | Deposit date: | 2013-07-24 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of HIV CRF07 B'/C gp41 reveals a hyper-mutant site in the middle of HR2 heptad repeat
Virology, 446, 2013
|
|
7BV5
 
 | | Crystal structure of the yeast heterodimeric ADAT2/3 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase subunit TAD2, tRNA-specific adenosine deaminase subunit TAD3 | | Authors: | Xie, W, Liu, X, Chen, R, Sun, Y, Chen, R, Zhou, J, Tian, Q. | | Deposit date: | 2020-04-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the yeast heterodimeric ADAT2/3 deaminase.
Bmc Biol., 18, 2020
|
|
3PFT
 
 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
8Y7Y
 
 | | Local structure of HCoV-HKU1A spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y, Liu, X, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
4X0V
 
 | | Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 | | Descriptor: | Beta-1,3-1,4-glucanase | | Authors: | Meng, D, Liu, X, Wang, X, Li, F, Feng, Y. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structural Insights into the Substrate Specificity of a Glycoside Hydrolase Family 5 Lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 2017
|
|
5FID
 
 | |
5EGN
 
 | |
