8JEU
 
 | |
8JEC
 
 | |
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
2OZB
 
 | | Structure of a human Prp31-15.5K-U4 snRNA complex | | Descriptor: | CALCIUM ION, RNA comprising the 5' Stem-Loop RNA of U4snRNA, U4/U6 small nuclear ribonucleoprotein Prp31, ... | | Authors: | Liu, S, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP.
Science, 316, 2007
|
|
4KBK
 
 | |
4KBA
 
 | | CK1d in complex with 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine inhibitor | | Descriptor: | 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine, Casein kinase I isoform delta, SULFATE ION | | Authors: | Liu, S. | | Deposit date: | 2013-04-23 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ligand-protein interactions of selective casein kinase 1 delta inhibitors.
J.Med.Chem., 56, 2013
|
|
4KBC
 
 | |
4KB8
 
 | |
8CUH
 
 | | Crystal structure of human TEAD2 complexed with its inhibitor TM2. | | Descriptor: | 4-[3-(2-cyclohexylethoxy)benzoyl]-N-phenylpiperazine-1-carboxamide, Transcriptional enhancer factor TEF-4 | | Authors: | Liu, S, Luo, X. | | Deposit date: | 2022-05-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a new class of reversible TEA-domain transcription factor inhibitors with a novel binding mode.
Elife, 11, 2022
|
|
4YHV
 
 | | Yeast Prp3 C-terminal fragment 325-469 | | Descriptor: | ACETIC ACID, U4/U6 small nuclear ribonucleoprotein PRP3 | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
4YHW
 
 | | Yeast Prp3 (296-469) in complex with fragment of U4/U6 di-snRNA | | Descriptor: | U4 snRNA fragment, U4/U6 small nuclear ribonucleoprotein PRP3, U6 snRNA fragment | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
1OQF
 
 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
8FBE
 
 | | Crystal structure of OrfX1 from Clostridium botulinum E1 | | Descriptor: | Neurotoxin complex component Orf-X1 | | Authors: | Liu, S, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
8FBD
 
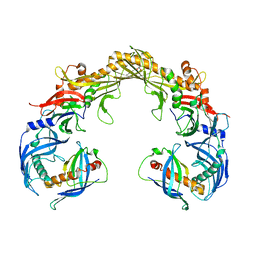 | | Crystal structure of OrfX1-OrfX3 complex from Clostridium botulinum E1 | | Descriptor: | ACETATE ION, Neurotoxin complex component Orf-X1, Neurotoxin complex component Orf-X3 | | Authors: | Liu, S, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
8EQB
 
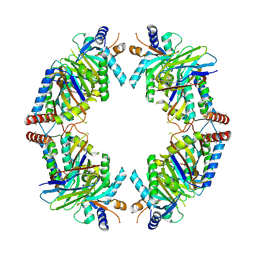 | | FAM46C/BCCIPalpha/Nanobody complex | | Descriptor: | Isoform 2 of BRCA2 and CDKN1A-interacting protein, Synthetic nanobody 1, Terminal nucleotidyltransferase 5C | | Authors: | Liu, S, Chen, H, Yin, Y, Bai, X, Zhang, X. | | Deposit date: | 2022-10-07 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of FAM46/TENT5 activity by BCCIP alpha adopting a unique fold.
Sci Adv, 9, 2023
|
|
8EXF
 
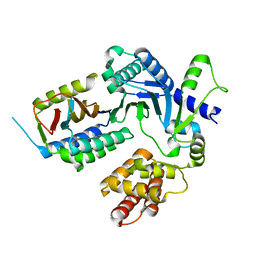 | |
8EXE
 
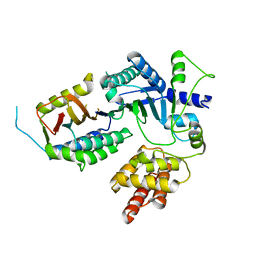 | |
1S2W
 
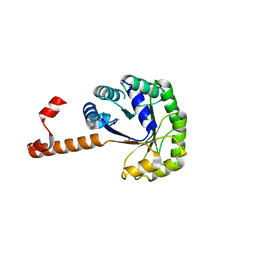 | | Crystal structure of phosphoenolpyruvate mutase in high ionic strength | | Descriptor: | Phosphoenolpyruvate phosphomutase, SULFATE ION | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2T
 
 | | Crystal Structure Of Apo Phosphoenolpyruvate Mutase | | Descriptor: | Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2U
 
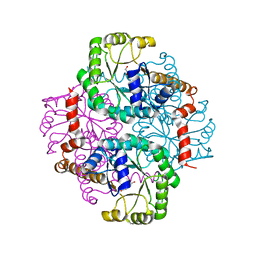 | | Crystal structure of the D58A phosphoenolpyruvate mutase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2V
 
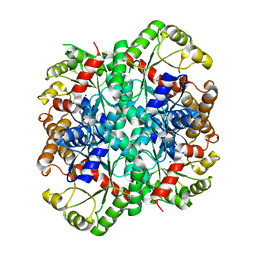 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | Descriptor: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
4DOY
 
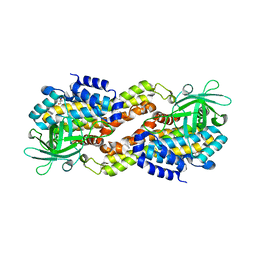 | | Crystal structure of Dibenzothiophene desulfurization enzyme C | | Descriptor: | Dibenzothiophene desulfurization enzyme C, GLYCEROL | | Authors: | Liu, S, Zhang, C, Zhu, D, Gu, L. | | Deposit date: | 2012-02-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of DszC from Rhodococcus sp. XP at 1.79 angstrom
Proteins, 82, 2014
|
|
3S41
 
 | | Glucokinase in complex with activator and glucose | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2011-05-18 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Designing glucokinase activators with reduced hypoglycemia risk: discovery of N,N-dimethyl-5-(2-methyl-6-((5-methylpyrazin-2-yl)-carbamoyl)benzofuran-4-yloxy)pyrimidine-2-carboxamide as a clinical candidate for the treatment of type 2 diabetes mellitus
MEDCHEMCOMM, 2, 2011
|
|
4LU9
 
 | | Crystal structure of E.coli SbcD at 2.5 angstrom resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1XG4
 
 | | Crystal Structure of the C123S 2-Methylisocitrate Lyase Mutant from Escherichia coli in complex with the inhibitor isocitrate | | Descriptor: | ISOCITRIC ACID, MAGNESIUM ION, Probable methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Han, Y, Melamud, E, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of 2-Methylisocitrate Lyase in Complex with Product and with Isocitrate Inhibitor Provide Insight into Lyase Substrate Specificity, Catalysis and Evolution
Biochemistry, 44, 2005
|
|
