7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
3PFT
 
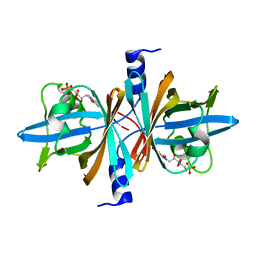 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
4ROV
 
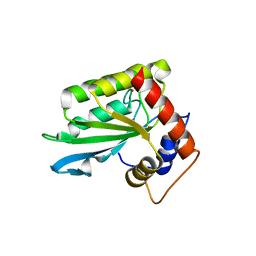 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
4ROW
 
 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
6THY
 
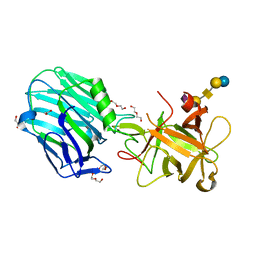 | | Botulinum neurotoxin A3 Hc domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BoNT/A3, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2019-11-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of botulinum neurotoxin subtype A3 cell binding domain in complex with GD1a co-receptor ganglioside.
Febs Open Bio, 10, 2020
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6ZNS
 
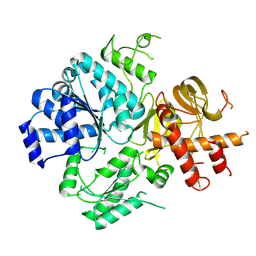 | | Crystal Structure of DUF1998 helicase MrfA | | Descriptor: | Uncharacterized ATP-dependent helicase YprA, ZINC ION | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
6ZNP
 
 | | Crystal Structure of DUF1998 helicase MrfA bound to DNA | | Descriptor: | CITRIC ACID, Uncharacterized ATP-dependent helicase YprA, ZINC ION, ... | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
6ZNQ
 
 | | Crystal Structure of DUF1998 helicase MrfA bound to DNA and AMPPNP | | Descriptor: | CITRIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized ATP-dependent helicase YprA, ... | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
5JCD
 
 | | Crystal structure of OsCEBiP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor-binding protein | | Authors: | Chai, J.J, Liu, S.M, Wang, J.Z. | | Deposit date: | 2016-04-15 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP
Structure, 24, 2016
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6WJJ
 
 | | Anthrax octamer prechannel bound to full-length lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen, ... | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
5GZR
 
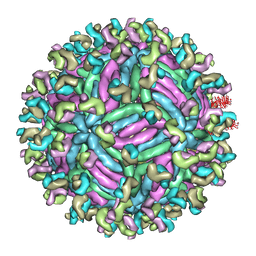 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
1B89
 
 | | CLATHRIN HEAVY CHAIN PROXIMAL LEG SEGMENT (BOVINE) | | Descriptor: | PROTEIN (CLATHRIN HEAVY CHAIN) | | Authors: | Ybe, J.A, Brodsky, F.M, Hofmann, K, Lin, K, Liu, S.-H, Chen, L, Earnest, T.N, Fletterick, R.J, Hwang, P.K. | | Deposit date: | 1999-05-27 | | Release date: | 1999-06-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Clathrin self-assembly is mediated by a tandemly repeated superhelix.
Nature, 399, 1999
|
|
6PSN
 
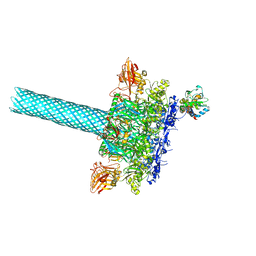 | | Anthrax toxin protective antigen channels bound to lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
5HRA
 
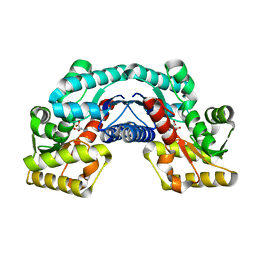 | | Crystal structure of an aspartate/glutamate racemase in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
1MCT
 
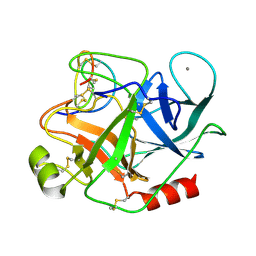 | | THE REFINED 1.6 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PORCINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, TRYPSIN INHIBITOR A | | Authors: | Huang, Q, Liu, S, Tang, Y. | | Deposit date: | 1992-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined 1.6 A resolution crystal structure of the complex formed between porcine beta-trypsin and MCTI-A, a trypsin inhibitor of the squash family. Detailed comparison with bovine beta-trypsin and its complex.
J.Mol.Biol., 229, 1993
|
|
1EPT
 
 | | REFINED 1.8 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF PORCINE EPSILON-TRYPSIN | | Descriptor: | CALCIUM ION, PORCINE E-TRYPSIN | | Authors: | Huang, Q, Wang, Z, Li, Y, Liu, S, Tang, Y. | | Deposit date: | 1994-06-07 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A resolution crystal structure of the porcine epsilon-trypsin.
Biochim.Biophys.Acta, 1209, 1994
|
|
4WHW
 
 | | Direct photocapture of bromodomains using tropolone chemical probes | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-{1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazol-5-yl}cyclohepta-2,4,6-trien-1-one, Bromodomain-containing protein 4 | | Authors: | Hett, E.C, Piatnitski Chekler, E.L, Basak, A, Bonin, P.D, Denny, R.A, Flick, A.C, Geoghegan, K.F, Liu, S, Pletcher, M.T, Robinson, R.P, Sahasrabudhe, P, Salter, S, Stock, I.A, Jones, L.H. | | Deposit date: | 2014-09-23 | | Release date: | 2015-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Direct photocapture of bromodomains using tropolone chemical probes
To Be Published
|
|
4WXM
 
 | | FleQ REC domain from Pseudomonas aeruginosa PAO1 | | Descriptor: | Transcriptional regulator FleQ | | Authors: | Su, T, Liu, S, Gu, L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The REC domain mediated dimerization is critical for FleQ from Pseudomonas aeruginosa to function as a c-di-GMP receptor and flagella gene regulator
J.Struct.Biol., 192, 2015
|
|
5HRC
 
 | | Crystal structure of an aspartate/glutamate racemase in complex with L-aspartate | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ASPARTIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
5HQT
 
 | | Crystal structure of an aspartate/glutamate racemase from Escherichia coli O157 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
6OIT
 
 | | CryoEM structure of Arabidopsis DDR' complex (DRD1 peptide-DMS3-RDM1) | | Descriptor: | Protein CHROMATIN REMODELING 35, Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | Authors: | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
6OIS
 
 | | CryoEM structure of Arabidopsis DR complex (DMS3-RDM1) | | Descriptor: | Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | Authors: | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
5GT2
 
 | | Crystal Structure and Biochemical Features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable deferrochelatase/peroxidase YfeX | | Authors: | Ma, Y.L, Yuan, Z.G, Liu, S, Wang, J.X, Gu, L.C, Liu, X.H. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates
Biochem. Biophys. Res. Commun., 484, 2017
|
|
