5BWO
 
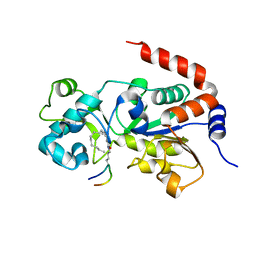 | | Crystal Structure of Human SIRT3 in Complex with a Palmitoyl H3K9 Peptide | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, PALMITIC ACID, ... | | Authors: | Gai, W, Jiang, H, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
5UBQ
 
 | | Cryo-EM structure of ciliary microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-21 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
5BWL
 
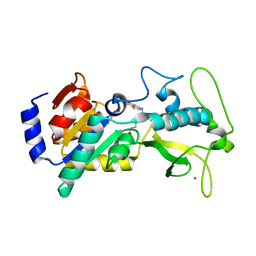 | |
5UCY
 
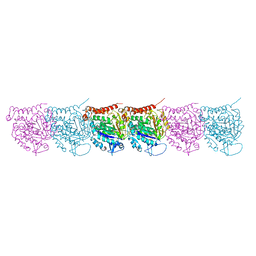 | | Cryo-EM map of protofilament of microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-22 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
5W5R
 
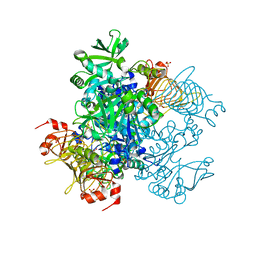 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase P96A mutant bound to activator pyruvate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, PYRUVIC ACID, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5W5T
 
 | | Agrobacterium tumefaciens ADP-Glucose Pyrophosphorylase bound to activator ethyl pyruvate | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION, ... | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5W6J
 
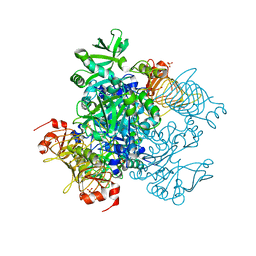 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Wu, R, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
7R6N
 
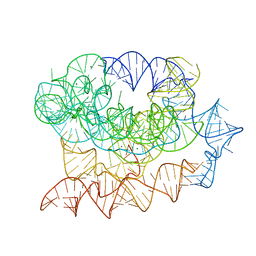 | | Exon-free state of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic trimeric construct | | Descriptor: | Group I intron, MAGNESIUM ION | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
7R6M
 
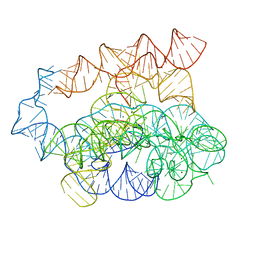 | | Post-2S intermediate of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic dimeric construct | | Descriptor: | Group I intron, Ligated exon mimic of the Group I intron, MAGNESIUM ION | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
7R6L
 
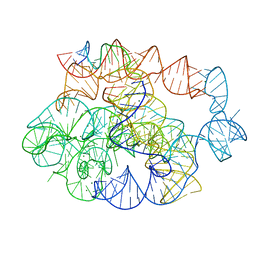 | | 5 prime exon-free pre-2S intermediate of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic dimeric construct | | Descriptor: | Group I intron, 3 prime fragment plus 3 prime exon, 5 prime fragment, ... | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
6M8H
 
 | | Crystal Structure of the R208Q mutant of G(i) subunit alpha-1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION | | Authors: | Mascarenhas, R, Goossens, J, Leverson, B, Kothawala, S, Ballicora, M, Olsen, K, de freitas, D, Liu, D. | | Deposit date: | 2018-08-21 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | FUNCTIONAL CONSEQUENCES OF ONCOGENIC MUTATIONS IN THE SWITCH II REGION OF Galphai1 and Galphas PROTEINS
To Be Published
|
|
7S5F
 
 | | Crystal structure of mannose-6-phosphate reductase from celery (Apium graveolens) leaves with NADP+ and mannonic acid bound | | Descriptor: | D-MANNONIC ACID, Manose-6-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
7S5I
 
 | | Crystal structure of Aldose-6-phosphate reductase (Ald6PRase) from peach (Prunus persica) leaves | | Descriptor: | Sorbitol-6-phosphate dehydrogenase | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
7L5F
 
 | |
7JH4
 
 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
6BY7
 
 | | Folding DNA into a lipid-conjugated nano-barrel for controlled reconstitution of membrane proteins | | Descriptor: | DNA (26-MER), DNA (27-MER), DNA (29-MER), ... | | Authors: | Dong, Y, Chen, S, Zhang, S, Sodroski, J, Yang, Z, Liu, D, Mao, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Folding DNA into a Lipid-Conjugated Nanobarrel for Controlled Reconstitution of Membrane Proteins.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7JX9
 
 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
7LON
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (1R,3S,4R)-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-methylcyclohexane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LOM
 
 | | Ornithine Aminotransferase (OAT) soaked with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (3~{S},4~{S})-4-methyl-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, (4~{R})-4-(fluoranylmethyl)-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
4MGR
 
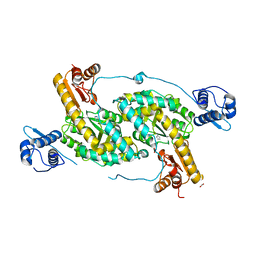 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2HR9
 
 | |
8V9M
 
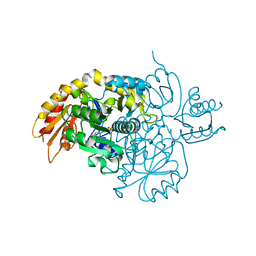 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (R)-3-amino-5,5-difluorocyclohex-1-ene-1-carboxylic acid. | | Descriptor: | 3-fluoro-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]benzoic acid, GLYCEROL, Ornithine aminotransferase, ... | | Authors: | Vargas, A.L, Devitt, A, Kaley, N, Silverman, R, Liu, D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, and Mechanistic Studies of ( R )-3-Amino-5,5-difluorocyclohex-1-ene-1-carboxylic Acid as an Inactivator of Human Ornithine Aminotransferase.
Acs Chem.Biol., 19, 2024
|
|
2GS4
 
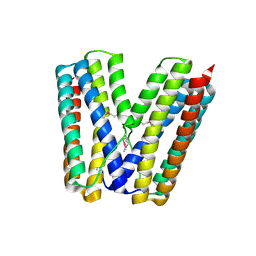 | | The crystal structure of the E.coli stress protein YciF. | | Descriptor: | Protein yciF | | Authors: | Hindupur, A, Liu, D, Zhao, Y, Bellamy, H.D, White, M.A, Fox, R.O. | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the E. coli stress protein YciF.
Protein Sci., 15, 2006
|
|
2JQT
 
 | | Structure of the bacterial replication origin-associated protein Cnu | | Descriptor: | H-NS/stpA-binding protein 2 | | Authors: | Bae, S.H, Liu, D, Lim, H.M, Lee, Y, Choi, B.S. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the nucleoid-associated protein Cnu reveals common binding sites for H-NS in Cnu and Hha.
Biochemistry, 47, 2008
|
|
2K6B
 
 | |
