5BMM
 
 | | Src in complex with DNA-templated macrocyclic inhibitor MC25b | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, macrocyclic inhibitor MC25b | | Authors: | Georghiou, G, Guja, K.E, Aleem, S, Kleiner, R.E, Liu, D.R, Miller, W.T, Garcia-Diaz, M, Seeliger, M.A. | | Deposit date: | 2015-05-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Basis for Intracellular Kinase Inhibition by Src-specific Peptidic Macrocycles.
Cell Chem Biol, 23, 2016
|
|
7LNM
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclopentene-1-carboxylic acid | | Descriptor: | (1~{R},3~{S},4~{R})-3-methyl-4-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Catlin, D, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LON
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (1R,3S,4R)-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-methylcyclohexane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LOM
 
 | | Ornithine Aminotransferase (OAT) soaked with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (3~{S},4~{S})-4-methyl-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, (4~{R})-4-(fluoranylmethyl)-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
5BKM
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-20 | | Release date: | 2022-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine)
To Be Published
|
|
2JQT
 
 | | Structure of the bacterial replication origin-associated protein Cnu | | Descriptor: | H-NS/stpA-binding protein 2 | | Authors: | Bae, S.H, Liu, D, Lim, H.M, Lee, Y, Choi, B.S. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the nucleoid-associated protein Cnu reveals common binding sites for H-NS in Cnu and Hha.
Biochemistry, 47, 2008
|
|
2K6B
 
 | |
2HR9
 
 | |
8F61
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine Quasi-Anaerobically | | Descriptor: | (5S)-5-methyl-1,3-diazinane-2,4-dione, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
4ZSW
 
 | | Pig Brain GABA-AT inactivated by (E)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
7JH4
 
 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
7LJT
 
 | | Porcine Dihydropyrimidine Dehydrogenase (DPD) soaked with 5-Ethynyluracil (5EU), NADPH - 20 minutes | | Descriptor: | 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7LJS
 
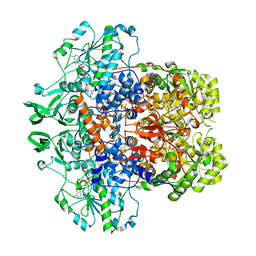 | | Porcine Dihydropyrimidine dehydrogenase (DPD) complexed with 5-Ethynyluracil (5EU) - Open Form | | Descriptor: | 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7LJU
 
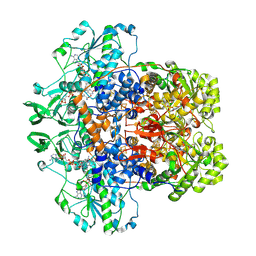 | | Porcine Dihydropyrimidine Dehydrogenase (DPD) crosslinked with 5-Ethynyluracil (5EU) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
9D6J
 
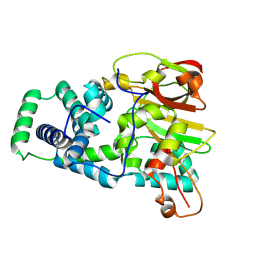 | | Nitrile hydratase BR52K mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
9D6M
 
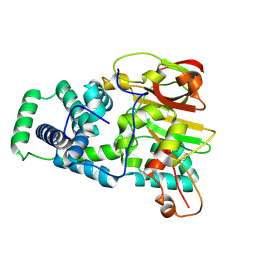 | | Nitrile hydratase BR157K mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
9D65
 
 | | Nitrile hydratase BR52A mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
4Y0I
 
 | | GABA-aminotransferase inactivated by conformationally-restricted inactivator | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Sanishvili, R, Lee, H.V, Dustin, D.H, Emma, D, Neil, K, Silverman, R.B, Liu, D. | | Deposit date: | 2015-02-06 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | GABA-aminotransferase inactivated by conformationally-restricted inactivator
To Be Published
|
|
9D6K
 
 | | Nitrile hydratase BR157A mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, OXYGEN ATOM | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-15 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
5BWN
 
 | | Crystal Structure of SIRT3 with a H3K9 Peptide Containing a Myristoyl Lysine | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | Authors: | Gai, W, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
5BWO
 
 | | Crystal Structure of Human SIRT3 in Complex with a Palmitoyl H3K9 Peptide | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, PALMITIC ACID, ... | | Authors: | Gai, W, Jiang, H, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
5BWL
 
 | |
5UBL
 
 | | A circularly permuted version of PvdQ (cpPvdQ) | | Descriptor: | Acyl-homoserine lactone acylase PvdQ | | Authors: | Wu, R, Mascarenhas, R, Catlin, D, Clevenger, K, Fast, W, Liu, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-01 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Circular Permutation Reveals a Chromophore Precursor Binding Pocket of the Siderophore Tailoring Enzyme PvdQ
To Be Published
|
|
8F5W
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine and NADPH Quasi-Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
4ZGJ
 
 | | Double Mutant H80A/H81A of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-23 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
