7PY9
 
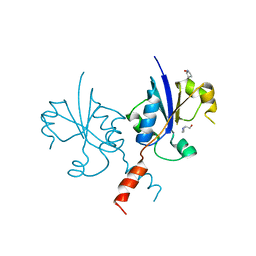 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with (pyrrolidin-2-yl)methanol | | Descriptor: | Cholinephosphate cytidylyltransferase, D-Prolinol | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-09 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7PYB
 
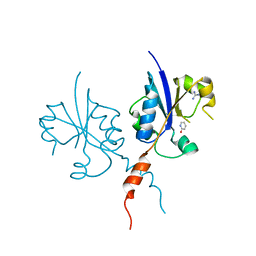 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 2-Hydroxypyridine | | Descriptor: | 1~{H}-pyridin-2-one, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-09 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7PYC
 
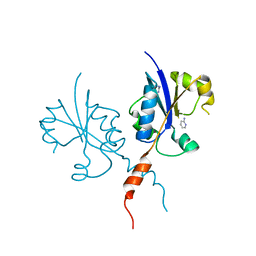 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 2-Aminopyridine | | Descriptor: | 2-AMINOPYRIDINE, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-09 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q9W
 
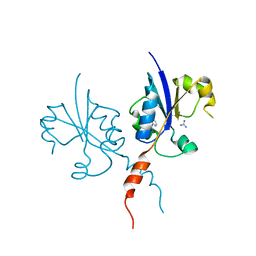 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 4-(aminomethyl)pyridin-2-amine | | Descriptor: | 4-(aminomethyl)pyridin-2-amine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7QA7
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Cyclopropanemethylamine | | Descriptor: | Cholinephosphate cytidylyltransferase, cyclopropylmethanamine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q9V
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1-(1,3-oxazol-4-yl)methanamine | | Descriptor: | 1,3-oxazol-4-ylmethanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QAD
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1,4-Oxazepane hydrochloride | | Descriptor: | 1,4-oxazepane, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7QD3
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with morpholine | | Descriptor: | Cholinephosphate cytidylyltransferase, morpholine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QVO
 
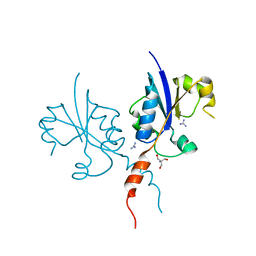 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with guanidine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QVN
 
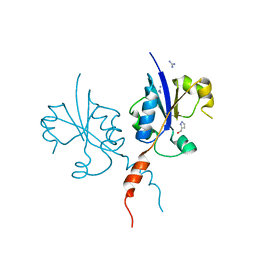 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with(1H-pyrazol-5-yl)methanol | | Descriptor: | 1~{H}-pyrazol-5-ylmethanol, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QBJ
 
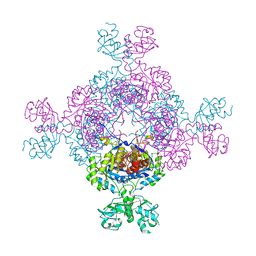 | | bacterial IMPDH chimera | | Descriptor: | Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7QDX
 
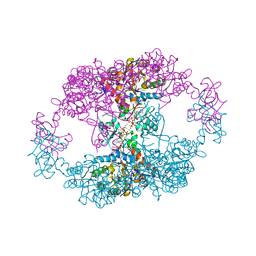 | | bacterial IMPDH chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7QEM
 
 | | bacterial IMPDH chimera | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Gedeon, A, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2021-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7R58
 
 | | Crystal structure of the GPVI-glenzocimab complex | | Descriptor: | CHLORIDE ION, Fab heavy chain, Fab light chain, ... | | Authors: | Jandrot-Perrus, M, Lebozec, K, Rose, N, Welin, M, Billiald, P. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Targeting platelet GPVI with glenzocimab: a novel mechanism for inhibition.
Blood Adv, 7, 2023
|
|
7SQO
 
 | | Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | McGrath, A.P, Kang, Y, Flinspach, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
6R29
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD cocrystallized with succinylphosphonate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R2A
 
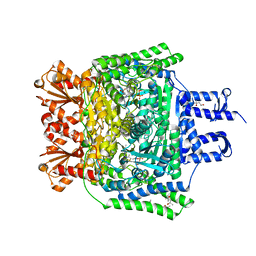 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD cocrystallized with succinylphosphonate phosphonoethyl ester (PESP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanyl-butanoic acid, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R2D
 
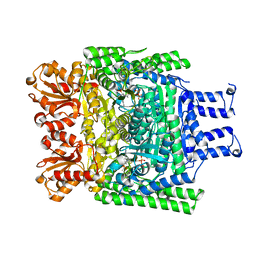 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate phosphonoethyl ester, followed by temperature increase | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanyl-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R2C
 
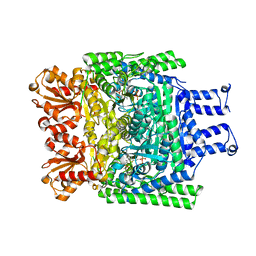 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate phosphonoethyl ester (PESP) | | Descriptor: | 4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanylidene-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R2B
 
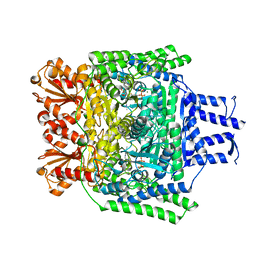 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
5EE7
 
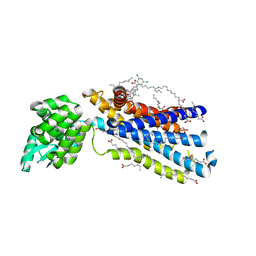 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
8P4Z
 
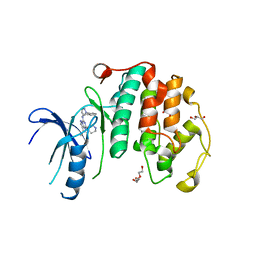 | | Crystal structure of the human CDK7 kinase domain in complex with LDC4297 | | Descriptor: | 2-[(3R)-piperidin-3-yl]oxy-8-propan-2-yl-N-[(2-pyrazol-1-ylphenyl)methyl]pyrazolo[1,5-a][1,3,5]triazin-4-amine, Cyclin-dependent kinase 7, GLYCEROL, ... | | Authors: | Laursen, M, Caing-Carlsson, R, Houssari, R, Javadi, A, Kimbung, Y.R, Murina, V, Orozco-Rodriguez, J.M, Svensson, A, Welin, M, Logan, D, Svensson, B, Walse, B. | | Deposit date: | 2023-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the human CDK7 kinase domain in complex with LDC4297
To Be Published
|
|
5H8L
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) C158S mutant in complex with putrescine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8J
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) in complex with cadaverine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
7TN9
 
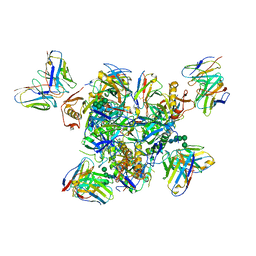 | | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, GP2, ... | | Authors: | Rayaprolu, V, Fulton, B, Rafique, A, Arturo, E, Williams, D, Hariharan, C, Callaway, H, Parvate, A, Schendel, S.L, Parekh, D, Hui, S, Shaffer, K, Pascal, K.E, Wloga, E, Giordano, S, Copin, R, Franklin, M, Boytz, R.M, Donahue, C, Davey, R, Baum, A, Kyratsous, C.A, Saphire, E.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the Inmazeb cocktail and resistance to Ebola virus escape.
Cell Host Microbe, 31, 2023
|
|
