3NEL
 
 | | Aspartyl-tRNA synthetase complexed with aspartic acid | | Descriptor: | ASPARTIC ACID, Aspartyl-tRNA synthetase | | Authors: | Schmitt, E, Moras, D, Moulinier, L. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation
Embo J., 17, 1998
|
|
3NEM
 
 | | Aspartyl-tRNA synthetase complexed with aspartyl adenylate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, Aspartyl-tRNA synthetase, ... | | Authors: | Schmitt, E, Moras, D, Moulinier, L. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation
Embo J., 17, 1998
|
|
3NEN
 
 | |
8G4A
 
 | |
6HQU
 
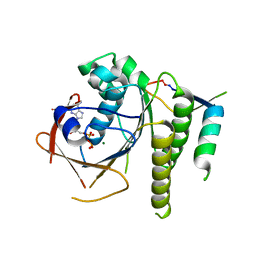 | | Humanised RadA mutant HumRadA22 in complex with a recombined BRC repeat 8-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Breast cancer type 2 susceptibility, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Lindenburg, L, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Improved RAD51 binders through motif shuffling based on the modularity of BRC repeats.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8K8T
 
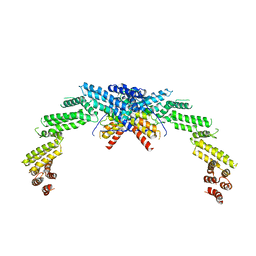 | | Structure of CUL3-RBX1-KLHL22 complex | | Descriptor: | Cullin-3, Kelch-like protein 22 | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
7QZP
 
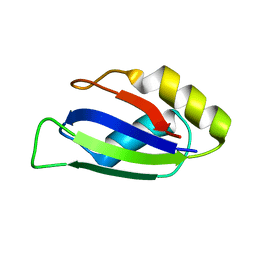 | | Identification and characterization of an RRM-containing, ELAV-like, RNA binding protein in Acinetobacter Baumannii | | Descriptor: | Hypothetical RNA binding protein from Acinetobacter baumannii | | Authors: | Ciani, C, Perez-Rafols, A, Bonomo, I, Micaelli, M, Esposito, A, Zucal, C, Belli, R, D'Agostino, V.G, Bianconi, I, Calderone, V, Cerofolini, L, Fragai, M, Provenzani, A. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii .
Biomolecules, 12, 2022
|
|
4KSI
 
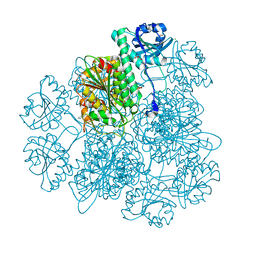 | | Crystal Structure Analysis of the Acidic Leucine Aminopeptidase of Tomato | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DuPrez, K.T, Scranton, M, Walling, L, Fan, L. | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of tomato wound-induced leucine aminopeptidase sheds light on substrate specificity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8SJK
 
 | | Pembrolizumab Caffeine crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY HEAVY CHAIN, ANTIBODY LIGHT CHAIN, ... | | Authors: | Larpent, P, Codan, L, Bothe, J.R, Stueber, D, Reichert, P, Fischmann, T, Su, Y, Pabit, S, Gupta, S, Iuzzolino, L, Cote, A. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Small-Angle X-ray Scattering as a Powerful Tool for Phase and Crystallinity Assessment of Monoclonal Antibody Crystallites in Support of Batch Crystallization.
Mol Pharm., 2024
|
|
1VLX
 
 | | STRUCTURE OF ELECTRON TRANSFER (COBALT-PROTEIN) | | Descriptor: | AZURIN, COBALT (II) ION | | Authors: | Bonander, N, Vanngard, T, Tsai, L.-C, Langer, V, Nar, H, Sjolin, L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The metal site of Pseudomonas aeruginosa azurin, revealed by a crystal structure determination of the Co(II) derivative and Co-EPR spectroscopy.
Proteins, 27, 1997
|
|
1MZZ
 
 | | Crystal Structure of Mutant (M182T)of Nitrite Reductase | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Guo, H, Olesen, K, Xue, Y, Shapliegh, J, Sjolin, L. | | Deposit date: | 2002-10-10 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The High resolution Crystal Structures of Nitrite Reductase and its mutant Met182Thr from Rhodobacter Sphaeroides Reveal a Gating Mechanism for the Electron Transfer to the Type 1 Copper Center
To be Published
|
|
1MZY
 
 | | Crystal Structure of Nitrite Reductase | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MAGNESIUM ION | | Authors: | Guo, H, Olesen, K, Xue, Y, Shapliegh, J, Sjolin, L. | | Deposit date: | 2002-10-10 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The High resolution Crystal Structures of Nitrite Reductase and its mutant Met182Thr from Rhodobacter Sphaeroides Reveal a Gating Mechanism for the Electron Transfer to the Type 1 Copper Center
To be Published
|
|
1N70
 
 | |
7SFA
 
 | |
7SF9
 
 | |
8OGD
 
 | | Structure of zinc(II) double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, THIOCYANATE ION, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
8OGE
 
 | | Structure of cobalt(II) substituted double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, COBALT (II) ION, Carbonic anhydrase 2, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
1XB3
 
 | | The D62C/K74C double mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
1TJM
 
 | | Crystallographic Identification of Sr2+ Coordination Site in Synaptotagmin I C2B Domain | | Descriptor: | GLYCEROL, STRONTIUM ION, Synaptotagmin I | | Authors: | Cheng, Y, Sequeira, S.M, Malinina, L, Tereshko, V, Sollner, T.H, Patel, D.J. | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain
Protein Sci., 13, 2004
|
|
1XB8
 
 | | Zn substituted form of D62C/K74C double mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, ZINC ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
1XB6
 
 | | The K24R mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
2VA1
 
 | | Crystal structure of UMP kinase from Ureaplasma parvum | | Descriptor: | PHOSPHATE ION, URIDYLATE KINASE | | Authors: | Egeblad-Welin, L, Welin, M, Wang, L, Eriksson, S. | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Investigations of Ureaplasma Parvum Ump Kinase - a Potential Antibacterial Drug Target
FEBS J., 274, 2007
|
|
4J2I
 
 | | Multiple crystal structures of an all-AT DNA dodecamer stabilized by weak interactions | | Descriptor: | 5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3' | | Authors: | Acosta-Reyes, F.J, Subirana, J.A, Pous, J, Sanchez, R, Condom, N, Baldini, R, Malinina, L, Campos, J.L. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
4J39
 
 | |
