7Q3H
 
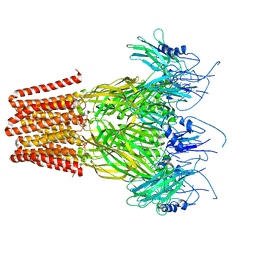 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM EDTA | | Descriptor: | Neur_chan_LBD domain-containing protein | | Authors: | Lycksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q3G
 
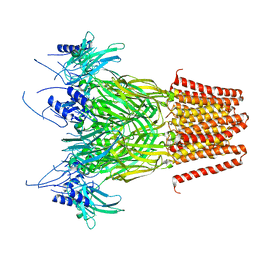 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM Ca2+ | | Descriptor: | CALCIUM ION, Neur_chan_LBD domain-containing protein | | Authors: | Licksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QWS
 
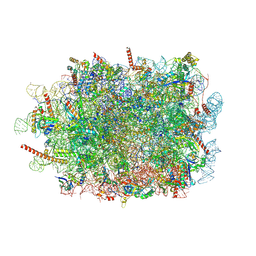 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
6T8H
 
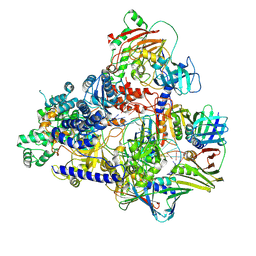 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
7QWQ
 
 | | Ternary complex of ribosome nascent chain with SRP and NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
8OWX
 
 | | Crystal Structure of METTL6 bound to SAH | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7QWR
 
 | | Structure of the ribosome-nascent chain containing an ER signal sequence in complex with NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
5LFF
 
 | | NMR structure of peptide 2 targeting CXCR4 | | Descriptor: | ARG-ALA-CYS-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
6RFE
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 4 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
7CJW
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a crowded environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
3HO0
 
 | | Crystal structure of the PPARgamma-LBD complexed with a new aryloxy-3phenylpropanoic acid | | Descriptor: | (2S)-2-(4-phenethylphenoxy)-3-phenyl-propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Laghezza, A, Lavecchia, A, Novellino, E. | | Deposit date: | 2009-06-01 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New 2-Aryloxy-3-phenyl-propanoic Acids As Peroxisome Proliferator-Activated Receptors alpha/gamma Dual Agonists with Improved Potency and Reduced Adverse Effects on Skeletal Muscle Function
J.Med.Chem., 52, 2009
|
|
3HOD
 
 | | Crystal structure of the PPARgamma-LBD complexed with a new aryloxy-3phenylpropanoic acid | | Descriptor: | (2S)-2-(4-benzylphenoxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Laghezza, A, Lavecchia, A, Novellino, E. | | Deposit date: | 2009-06-02 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New 2-Aryloxy-3-phenyl-propanoic Acids As Peroxisome Proliferator-Activated Receptors alpha/gamma Dual Agonists with Improved Potency and Reduced Adverse Effects on Skeletal Muscle Function
J.Med.Chem., 52, 2009
|
|
7D8K
 
 | | Solution structure of the methyl-CpG binding domain of MBD6 from Arabidopsis thaliana | | Descriptor: | Methyl-CpG-binding domain-containing protein 6 | | Authors: | Mahana, Y, Ohki, I, Walinda, E, Morimoto, D, Sugase, K, Shirakawa, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Methylated DNA Recognition by the Methyl-CpG Binding Domain of MBD6 from Arabidopsis thaliana .
Acs Omega, 7, 2022
|
|
6RFF
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 7 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-nitro-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
5CXZ
 
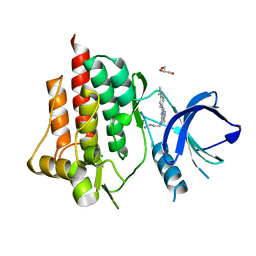 | | SYK catalytic domain complexed with naphthyridine inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, N-{7-[4-(dimethylamino)phenyl]-1,6-naphthyridin-5-yl}propane-1,3-diamine, ... | | Authors: | Thoma, G, Veenstra, S, Strang, R, Blanz, J, Vangrevelinghe, E, Berghausen, J, Lee, C.C, Zerwes, H.-G. | | Deposit date: | 2015-07-29 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Orally bioavailable Syk inhibitors with activity in a rat PK/PD model.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6RCM
 
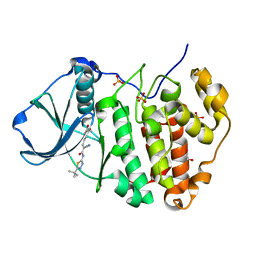 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 3 | | Descriptor: | (~{E})-~{N}-(5-~{tert}-butyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
7CJV
 
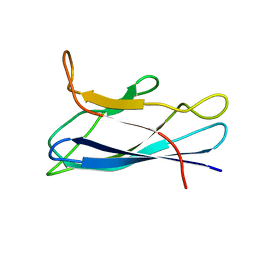 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a dilute environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
1W26
 
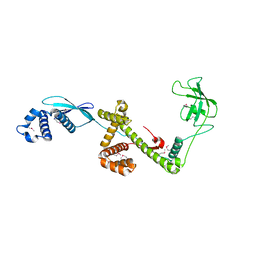 | | Trigger Factor in Complex with the Ribosome forms a Molecular Cradle for Nascent Proteins | | Descriptor: | TRIGGER FACTOR | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-06-28 | | Release date: | 2004-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
8QO4
 
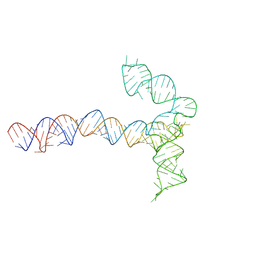 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | MERS-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO3
 
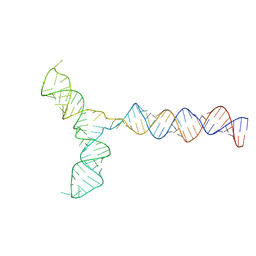 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | RoBat-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO5
 
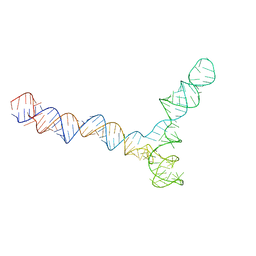 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | SARS-CoV-2-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO2
 
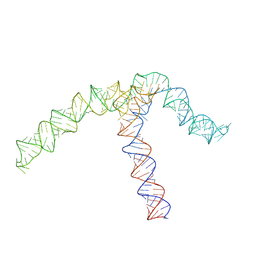 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | OC43-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8ATG
 
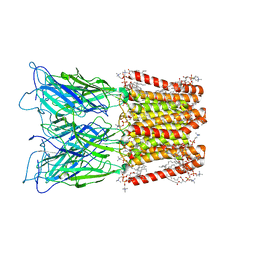 | | Pentameric ligand-gated ion channel GLIC with bound lipids | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Proton-gated ion channel | | Authors: | Bergh, C, Rovsnik, U, Howard, R.J, Lindahl, E. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of lipid binding sites in a ligand-gated ion channel by integrating simulations and cryo-EM.
Elife, 12, 2024
|
|
8BTK
 
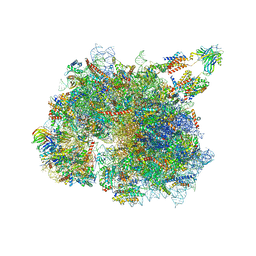 | | Structure of the TRAP complex with the Sec translocon and a translating ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jaskolowski, M, Jomaa, A, Gamerdinger, M, Shrestha, S, Leibundgut, M, Deuerling, E, Ban, N. | | Deposit date: | 2022-11-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of the TRAP complex function in ER protein biogenesis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8B7V
 
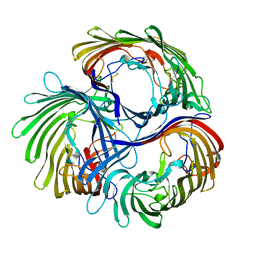 | | Automated simulation-based refinement of maltoporin into a cryo-EM density | | Descriptor: | Maltoporin | | Authors: | Yvonnesdotter, L, Rovsnik, U, Blau, C, Lycksell, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2022-10-03 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Automated simulation-based membrane protein refinement into cryo-EM data.
Biophys.J., 122, 2023
|
|
