1Z1I
 
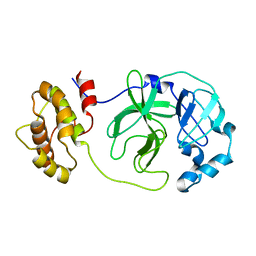 | | Crystal structure of native SARS CLpro | | Descriptor: | 3C-like proteinase | | Authors: | Liang, P.H, Wang, A.H. | | Deposit date: | 2005-03-04 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Understanding the maturation process and inhibitor design of SARS-CoV 3CLpro from the crystal structure of C145A in a product-bound form
J.Biol.Chem., 280, 2005
|
|
2YPW
 
 | | Atomic model for the N-terminus of TraO fitted in the full-length structure of the bacterial pKM101 type IV secretion system core complex | | Descriptor: | TRAO | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-02 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.4 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
7SR8
 
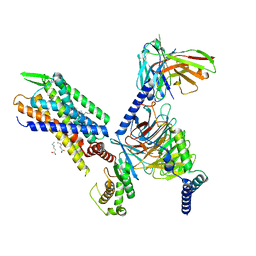 | | Molecular mechanism of the the wake-promoting agent TAK-925 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hypocretin receptor type 2, ... | | Authors: | Yin, J, Chapman, K, Lian, P, De Brabander, J.K, Rosenbaum, D.M. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
8EIT
 
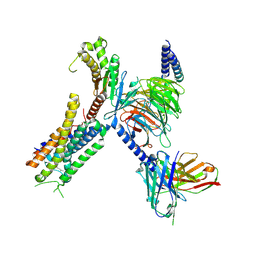 | | Structure of FFAR1-Gq complex bound to DHA | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-15 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJK
 
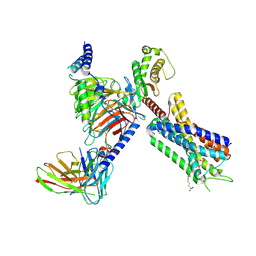 | | Structure of FFAR1-Gq complex bound to TAK-875 in a lipid nanodisc | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJC
 
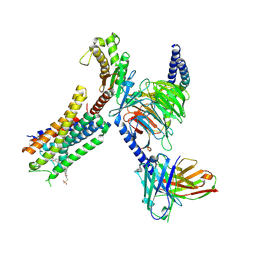 | | Structure of FFAR1-Gq complex bound to TAK-875 | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3ZBJ
 
 | | Fitting results in the I-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
4AG6
 
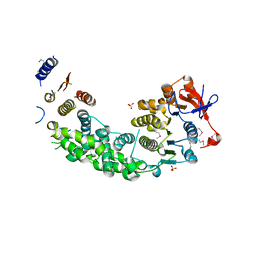 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | SULFATE ION, TYPE IV SECRETORY PATHWAY VIRB4 COMPONENTS-LIKE PROTEIN | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AG5
 
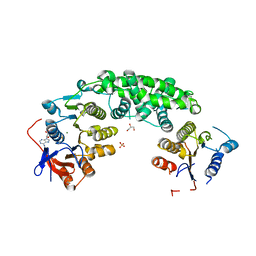 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZBI
 
 | | Fitting result in the O-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAF PROTEIN, TRAN PROTEIN, TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
1CX9
 
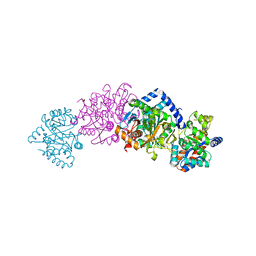 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-AMINOPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-AMINOPHENYLTHIO)-BUTYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-08-29 | | Release date: | 1999-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
2DH4
 
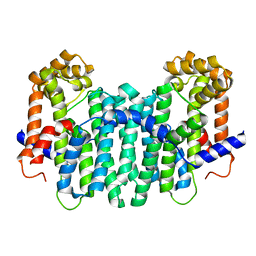 | | Geranylgeranyl pyrophosphate synthase | | Descriptor: | MAGNESIUM ION, YPL069C | | Authors: | Chang, T.-H, Guo, R.-T, Ko, T.-P, Wang, A.H, Liang, P.-H. | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of Type-III Geranylgeranyl Pyrophosphate Synthase from Saccharomyces cerevisiae and the Mechanism of Product Chain Length Determination.
J.Biol.Chem., 281, 2006
|
|
2DTN
 
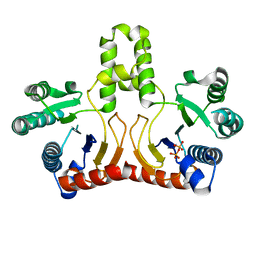 | | Crystal structure of Helicobacter pylori undecaprenyl pyrophosphate synthase complexed with pyrophosphate | | Descriptor: | DIPHOSPHATE, undecaprenyl pyrophosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chen, C.L, Ko, T.P, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2006-07-13 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical characterization, crystal structure, and inhibitors of Helicobacter pylori undecaprenyl pyrophosphate synthase
To be Published
|
|
1UEH
 
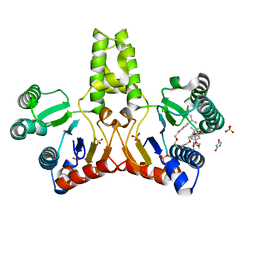 | | E. coli undecaprenyl pyrophosphate synthase in complex with Triton X-100, magnesium and sulfate | | Descriptor: | MAGNESIUM ION, OXTOXYNOL-10, SULFATE ION, ... | | Authors: | Chang, S.-Y, Ko, T.-P, Liang, P.-H, Wang, A.H.-J. | | Deposit date: | 2003-05-15 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton
J.Biol.Chem., 278, 2003
|
|
7C8T
 
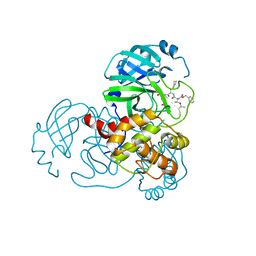 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0205221 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
7C8R
 
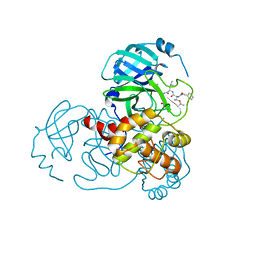 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0203770 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
4U3A
 
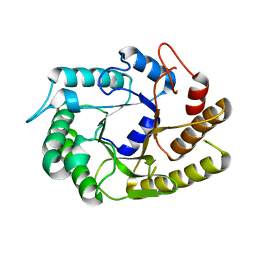 | | Crystal structure of CtCel5E | | Descriptor: | Endoglucanase H | | Authors: | Yuan, S.F, Liang, P.H, Ho, M.C. | | Deposit date: | 2014-07-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Biochemical Characterization and Structural Analysis of a Bifunctional Cellulase/Xylanase from Clostridium thermocellum
J.Biol.Chem., 290, 2015
|
|
2WSY
 
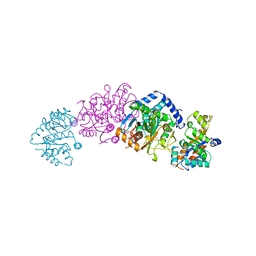 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
8EIZ
 
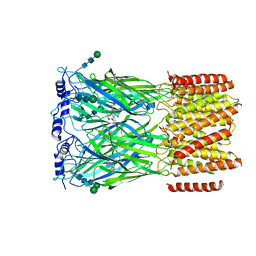 | | Cryo-EM structure of squid sensory receptor CRB1 | | Descriptor: | N-benzyl-2-(2,6-dimethylanilino)-N,N-diethyl-2-oxoethan-1-aminium, Squid sensory receptor CRB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, van Giesen, L, Kilian, P.B, Bai, X, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
1J4N
 
 | | Crystal Structure of the AQP1 water channel | | Descriptor: | AQUAPORIN 1, nonyl beta-D-glucopyranoside | | Authors: | Sui, H, Han, B.-G, Lee, J.K, Walian, P, Jap, B.K. | | Deposit date: | 2001-10-19 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of water-specific transport through the AQP1 water channel.
Nature, 414, 2001
|
|
3WJO
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli with isopentenyl pyrophosphate (IPP) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Octaprenyl diphosphate synthase | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-12 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
3WJK
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli | | Descriptor: | Octaprenyl diphosphate synthase | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-11 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
5BYW
 
 | |
3WJN
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli with farnesyl S-thiol-pyrophosphate (FSPP) | | Descriptor: | Octaprenyl diphosphate synthase, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-12 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
2AZL
 
 | | Crystal structure for the mutant F117E of Thermotoga maritima octaprenyl pyrophosphate synthase | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H. | | Deposit date: | 2005-09-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
