2PYE
 
 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry TCR Clone C5C1 Complexed with MHC | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Beta-2-microglobulin, Cancer/testis antigen 1B, ... | | Authors: | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | Deposit date: | 2007-05-16 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
1VEV
 
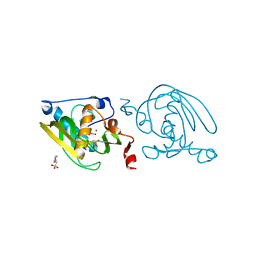 | | Crystal structure of peptide deformylase from Leptospira Interrogans (LiPDF) at pH6.5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMIC ACID, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
1VEY
 
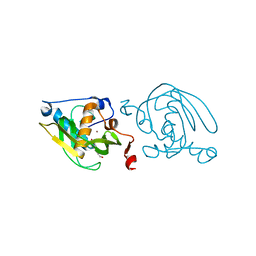 | | Crystal Structure of Peptide Deformylase from Leptospira Interrogans (LiPDF) at pH7.0 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peptide deformylase, ZINC ION | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
1VEZ
 
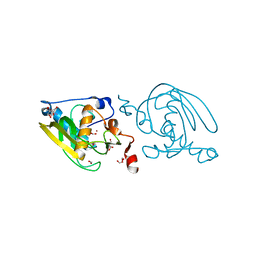 | | Crystal Structure of Peptide Deformylase from Leptospira Interrogans(LiPDF) at pH8.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
2JIV
 
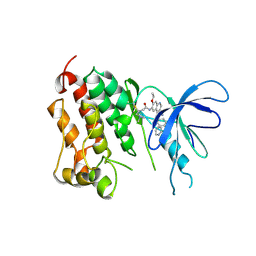 | | Crystal structure of EGFR kinase domain T790M mutation in compex with HKI-272 | | Descriptor: | CHLORIDE ION, EPIDERMAL GROWTH FACTOR RECEPTOR, N-(4-{[3-chloro-4-(pyridin-2-ylmethoxy)phenyl]amino}-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)butanamide | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Li, Y, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2KFV
 
 | | Structure of the amino-terminal domain of human FK506-binding protein 3 / Northeast Structural Genomics Consortium Target HT99A | | Descriptor: | FK506-binding protein 3 | | Authors: | Sunnerhagen, M, Davis, T, Gutmanas, A, Fares, C, Ouyang, H, Lemak, A, Li, Y, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C.H, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of FK506-binding protein 3
To be Published
|
|
7W3Q
 
 | | Crystal structure of RORgamma in complex with natural inverse agonist | | Descriptor: | (3S,5R,6S,8R,9R,10R,12R,13R,14R,17S)-4,4,8,10,14-pentamethyl-17-[(2R)-6-methyl-2-oxidanyl-hept-5-en-2-yl]-2,3,5,6,7,9,11,12,13,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthrene-3,6,12-triol, Nuclear receptor ROR-gamma, Peptide from Nuclear receptor coactivator 2 | | Authors: | Tian, S.Y, Li, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of RORgamma in complex with natural inverse agonist
To Be Published
|
|
7W3P
 
 | | Crystal structure of RORgamma in complex with natural inverse agonist | | Descriptor: | (3S,5R,8R,9R,10R,12R,13R,14R,17S)-4,4,8,10,14-pentamethyl-17-[(2R)-2,6,6-trimethyloxan-2-yl]-2,3,5,6,7,9,11,12,13,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthrene-3,12-diol, Nuclear receptor ROR-gamma, Peptide from Nuclear receptor coactivator 2 | | Authors: | Tian, S.Y, Li, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of RORgamma in complex with natural inverse agonist
To Be Published
|
|
3O6O
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of an the inhibitor BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein 83, PENTAETHYLENE GLYCOL | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Li, Y, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P, Fairlamb, A.H, Ferguson, M.A.J, Thompson, S, MacKenzie, C, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
3OMU
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of a thienopyrimidine derivative | | Descriptor: | 2-amino-4-{2,4-dichloro-5-[2-(diethylamino)ethoxy]phenyl}-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, Heat shock protein 83 | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Cossar, D, Li, Y, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-27 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
7MWI
 
 | | Crystal structure of human BAZ2A | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
3OPD
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of a benzamide derivative | | Descriptor: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-[(cis-4-hydroxycyclohexyl)amino]benzamide, Heat shock protein 83 | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Sullivan, H, Chamberlain, K, Weadge, J, Cossar, D, Li, Y, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
3QWP
 
 | | Crystal structure of SET and MYND domain containing 3; Zinc finger MYND domain-containing protein 1 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, SET and MYND domain-containing protein 3, ... | | Authors: | Dong, A, Dombrovski, L, Li, Y, Wernimont, A, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-28 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Crystal Structure of human Histone-lysine N-methyltransferase SMYD3
To be Published
|
|
6AOG
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF, and 5-(4-chlorophenyl)-6-ethylpyrimidine-2,4-diamine (pyrimethamine) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, ... | | Authors: | Thomas, S.B, Li, Y, Chen, Z, Lu, H. | | Deposit date: | 2017-08-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification of selective, brain penetrant Toxoplasma gondii dihydrofolate reductase inhibitors for the treatment of toxoplasmosis
To Be Published
|
|
6AOH
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF, and 5-(4-(3-(2-methoxypyrimidin-5-yl)phenyl)piperazin-1-yl)pyrimidine-2,4-diamine (TRC-2533) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-{4-[3-(2-methoxypyrimidin-5-yl)phenyl]piperazin-1-yl}pyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Li, Y, Chen, Z, Lu, H. | | Deposit date: | 2017-08-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification of selective, brain penetrant Toxoplasma gondii dihydrofolate reductase inhibitors for the treatment of toxoplasmosis
To Be Published
|
|
7XK1
 
 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (both in tRNA binding states) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK0
 
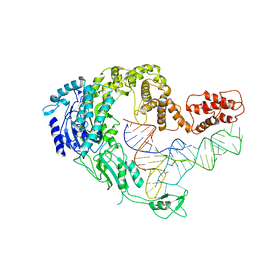 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA locked state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XV7
 
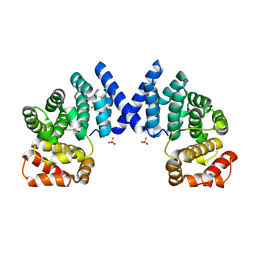 | |
7XJZ
 
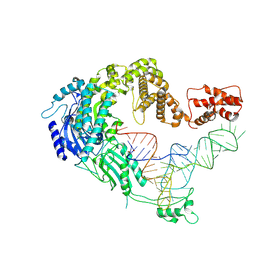 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA binding state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XJY
 
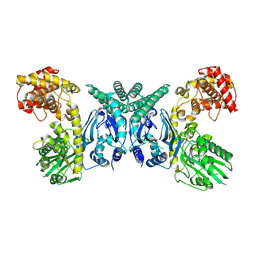 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase (apo form) | | Descriptor: | Glycine--tRNA ligase | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
6AOI
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF, and 5-(4-phenylpiperazin-1-yl)-6-propylpyrimidine-2,4-diamine (TRC-2528) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-phenylpiperazin-1-yl)-6-propylpyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Li, Y, Chen, Z, Lu, H. | | Deposit date: | 2017-08-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Identification of selective, brain penetrant Toxoplasma gondii dihydrofolate reductase inhibitors for the treatment of toxoplasmosis
To Be Published
|
|
7XRB
 
 | | human STK19 dimer | | Descriptor: | CHLORIDE ION, Isoform 2 of Serine/threonine-protein kinase 19, SULFATE ION | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2022-05-10 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | human STK19 dimer
To Be Published
|
|
7YX6
 
 | | Crystal structure of YTHDF2 with compound YLI_DF_027 | | Descriptor: | 6-ethyl-1H-pyrimidine-2,4-dione, GLYCEROL, SULFATE ION, ... | | Authors: | Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7YXE
 
 | | Crystal structure of YTHDF2 with compound ZA_143 | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Nai, F, Zalesak, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7Z26
 
 | | Crystal structure of YTHDF2 YTH domain in complex with m6A RNA | | Descriptor: | GLYCEROL, RNA (5'-R(P*(6MZ)P*CP*U)-3'), SULFATE ION, ... | | Authors: | Nai, F, Nachawati, R, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
