6AJ7
 
 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ3
 
 | | The structure of Enterovirus D68 procapsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
3SLI
 
 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2,7-ANHYDRO-NEU5AC PREPARED BY SOAKING WITH 3'-SIALYLLACTOSE | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
7R7T
 
 | | p47-bound p97-R155H mutant with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NSFL1 cofactor p47, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
8GTB
 
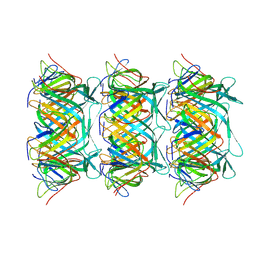 | | Cryo-EM structure of the marine siphophage vB_DshS-R4C tail tube protein | | Descriptor: | Major tail protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
3UIU
 
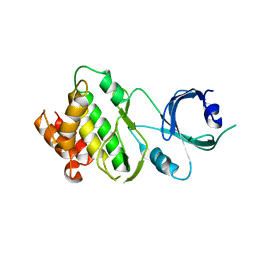 | | Crystal structure of Apo-PKR kinase domain | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
6V5C
 
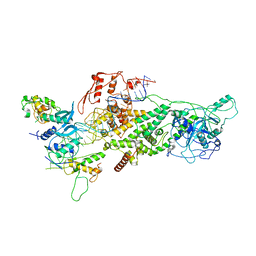 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | Descriptor: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
1MO0
 
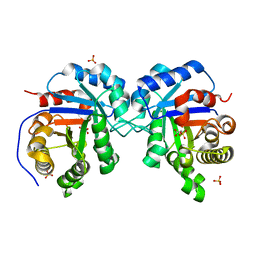 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | Descriptor: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6V5B
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - Active state | | Descriptor: | CALCIUM ION, Microprocessor complex subunit DGCR8, Pri-miR-16-2 (78-MER), ... | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
5YSW
 
 | | Crystal Structure Analysis of Rif16 in complex with R-L | | Descriptor: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5YSM
 
 | | Crystal Structure Analysis of Rif16 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
8GTF
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C stopper-terminator complex | | Descriptor: | Head-to-tail joining protein, Major tail protein, Terminator protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
6ONU
 
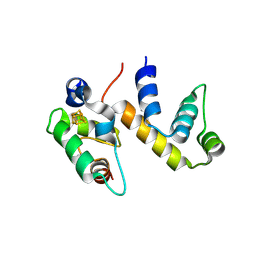 | | Complex structure of WhiB1 and region 4 of SigA in P21 space group. | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
6AD0
 
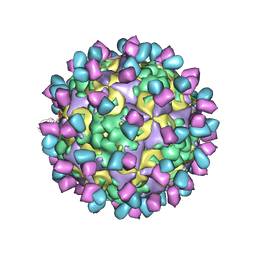 | | The structure of CVA10 mature virion in complex with Fab 2G8 | | Descriptor: | SPHINGOSINE, VH of Fab 2G8, VL of Fab 2G8, ... | | Authors: | Zhu, R, Zheng, Q.B, Xu, L.F, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
7PVN
 
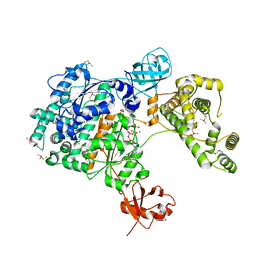 | | Crystal Structure of Human UBA6 in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Truongvan, N, Li, S, Schindelin, H. | | Deposit date: | 2021-10-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
7KUF
 
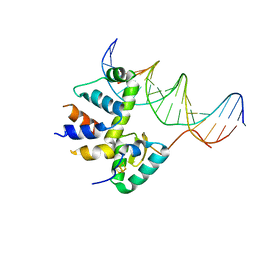 | | Transcription activation subcomplex with WhiB7 bound to SigmaAr4-RNAP Beta flap tip chimera and DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*AP*AP*AP*TP*CP*GP*GP*TP*TP*GP*TP*GP*GP*T)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*AP*AP*CP*CP*GP*AP*TP*TP*TP*TP*CP*T)-3'), IRON/SULFUR CLUSTER, ... | | Authors: | Wan, T, Horova, M, Beltran, D.G, Li, S.R, Zhang, L.M. | | Deposit date: | 2020-11-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the functional divergence of WhiB-like proteins in Mycobacterium tuberculosis.
Mol.Cell, 81, 2021
|
|
6ACZ
 
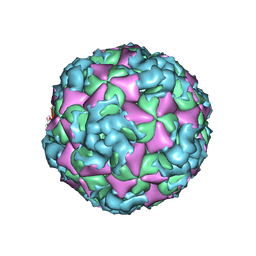 | | The structure of CVA10 virus A-particle from its complex with Fab 2G8 | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, R, Zheng, Q.B, Xu, L.F, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6ACY
 
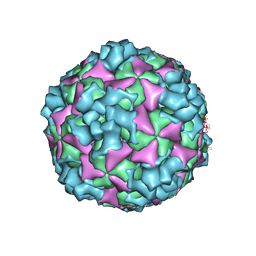 | | The structure of CVA10 virus A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Cui, Y.X, Zheng, Q.B, Zhu, R, Xu, L.F, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6IY1
 
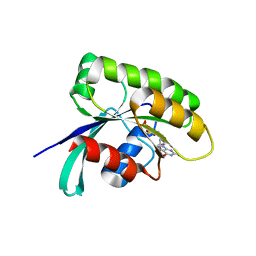 | | Structure of human Ras-related protein Rab11 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-11A | | Authors: | Ma, P, Li, S, Li, J. | | Deposit date: | 2018-12-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of human Ras-related protein Rab11
To Be Published
|
|
3MAZ
 
 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
8XEW
 
 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein | | Descriptor: | DSR2 H171A | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
8XFE
 
 | | Cryo-EM structure of defence-associated sirtuin 2 (DSR2) H171A protein in complex with DSR anti-defence 1(DSAD1) | | Descriptor: | DSAD1, DSR2(H171A) | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
6ONO
 
 | | Complex structure of WhiB1 and region 4 of SigA in C2221 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
