1BOZ
 
 | | STRUCTURE-BASED DESIGN AND SYNTHESIS OF LIPOPHILIC 2,4-DIAMINO-6-SUBSTITUTED QUINAZOLINES AND THEIR EVALUATION AS INHIBITORS OF DIHYDROFOLATE REDUCTASE AND POTENTIAL ANTITUMOR AGENTS | | Descriptor: | N6-(2,5-DIMETHOXY-BENZYL)-N6-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (DIHYDROFOLATE REDUCTASE) | | Authors: | Gangjee, A, Vidwans, A.P, Vasudevan, A, Queener, S.F, Kisliuk, R.L, Cody, V, Li, R, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1998-08-06 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation as inhibitors of dihydrofolate reductases and potential antitumor agents.
J.Med.Chem., 41, 1998
|
|
1EKM
 
 | | CRYSTAL STRUCTURE AT 2.5 A RESOLUTION OF ZINC-SUBSTITUTED COPPER AMINE OXIDASE OF HANSENULA POLYMORPHA EXPRESSED IN ESCHERICHIA COLI | | Descriptor: | COPPER AMINE OXIDASE, ZINC ION | | Authors: | Chen, Z, Schwartz, B, Williams, N.K, Li, R, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2000-03-09 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure at 2.5 A resolution of zinc-substituted copper amine oxidase of Hansenula polymorpha expressed in Escherichia coli.
Biochemistry, 39, 2000
|
|
5JFB
 
 | | Crystal structure of the scavenger receptor cysteine-rich domain 5 (SRCR5) from porcine CD163 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Ma, H, Jiang, L, Qiao, S, Zhang, G, Li, R. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection
J. Virol., 91, 2017
|
|
5Z65
 
 | | Crystal structure of porcine aminopeptidase N ectodomain in functional form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, Y.G, Li, R, Jiang, L.G, Qiao, S.L, Zhang, G.P. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Characterization of the interaction between recombinant porcine aminopeptidase N and spike glycoprotein of porcine epidemic diarrhea virus.
Int. J. Biol. Macromol., 117, 2018
|
|
4YR6
 
 | | Fab fragment of 5G6 in complex with epitope peptide | | Descriptor: | ACE-LYS-LEU-ARG-GLY-VAL-LEU-GLN-GLY-HIS-LEU, GLYCEROL, heavy chain of 5G6, ... | | Authors: | Tao, Y, Mo, X, Li, R. | | Deposit date: | 2015-03-14 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for the specific inhibition of glycoprotein Ib alpha shedding by an inhibitory antibody.
Sci Rep, 6, 2016
|
|
1EUJ
 
 | | A NOVEL ANTI-TUMOR CYTOKINE CONTAINS A RNA-BINDING MOTIF PRESENT IN AMINOACYL-TRNA SYNTHETASES | | Descriptor: | ENDOTHELIAL MONOCYTE ACTIVATING POLYPEPTIDE 2 | | Authors: | Kim, Y, Shin, J, Li, R, Cheong, C, Kim, S. | | Deposit date: | 2000-04-17 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel anti-tumor cytokine contains an RNA binding motif present in aminoacyl-tRNA synthetases.
J.Biol.Chem., 275, 2000
|
|
8K14
 
 | | X-ray crystal structure of 18a in BRD4(1) | | Descriptor: | 4-[8-methoxy-2-methyl-1-(1-phenylethyl)imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Xu, H, Shen, H, Zhang, Y, Xu, Y, Li, R. | | Deposit date: | 2023-07-10 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of (R)-4-(8-methoxy-2-methyl-1-(1-phenylethy)-1H-imidazo[4,5-c]quinnolin-7-yl)-3,5-dimethylisoxazole as a potent and selective BET inhibitor for treatment of acute myeloid leukemia (AML) guided by FEP calculation.
Eur.J.Med.Chem., 263, 2024
|
|
1M1U
 
 | | AN ISOLEUCINE-BASED ALLOSTERIC SWITCH CONTROLS AFFINITY AND SHAPE SHIFTING IN INTEGRIN CD11B A-DOMAIN | | Descriptor: | CALCIUM ION, Integrin alpha-M | | Authors: | Xiong, J.-P, Li, R, Essafi, M, Stehle, T, Arnaout, M.A. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An isoleucine-based allosteric switch controls affinity and shape shifting in integrin CD11b A-domain.
J.Biol.Chem., 275, 2000
|
|
8H7J
 
 | | The crystal structure of CD163 SRCR5-9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor cysteine-rich type 1 protein M130, ... | | Authors: | Luo, Z.P, Li, R, Ma, H.F. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of CD163 SRCR5-9
To Be Published
|
|
3GO6
 
 | | Crystal Structure of M. tuberculosis ribokinase (Rv2436) in complex with ribose and AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOKINASE RBSK, ... | | Authors: | Masters, E.I, Sun, Y, Wang, Y, Parker, W.B, Li, R. | | Deposit date: | 2009-03-18 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and biochemical characterization of M.tuberculosis ribokinase (Rv2436)
To be Published
|
|
5HRJ
 
 | |
3GO7
 
 | | Crystal Structure of M. tuberculosis ribokinase (Rv2436) in complex with ribose | | Descriptor: | MAGNESIUM ION, RIBOKINASE RBSK, alpha-D-ribofuranose | | Authors: | Masters, E.I, Sun, Y, Wang, Y, Parker, W.B, Li, R. | | Deposit date: | 2009-03-18 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and biochemical characterization of M.tuberculosis ribokinase (Rv2436)
To be Published
|
|
3RFE
 
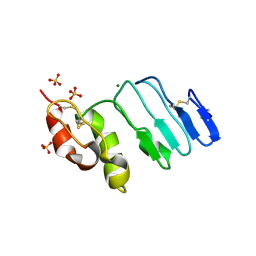 | | Crystal structure of glycoprotein GPIb ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-06 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.245 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIbbeta and a GPIbbeta/GPIX chimer
Blood, 118, 2011
|
|
3REZ
 
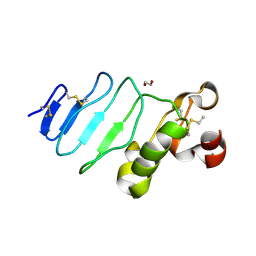 | | glycoprotein GPIb variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-05 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIb beta and a GPIb beta/GPIX chimera
Blood, 118, 2011
|
|
4EKU
 
 | |
4ISO
 
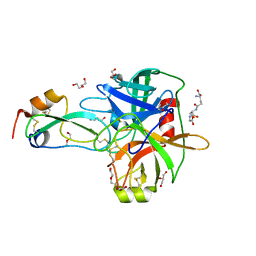 | | Crystal Structure of Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, GLYCEROL, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
4ISN
 
 | | Crystal Structure of Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | GLUTATHIONE, Kunitz-type protease inhibitor 1, Suppressor of tumorigenicity 14 protein, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
4IS5
 
 | | Crystal Structure of the ligand-free inactive Matriptase | | Descriptor: | GLUTATHIONE, GLYCEROL, SULFATE ION, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
4ISL
 
 | | Crystal Structure of the inactive Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | GLUTATHIONE, GLYCEROL, Kunitz-type protease inhibitor 1, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
6ITE
 
 | | Crystal structure of group A Streptococcal surface dehydrogenase (SDH) | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Yuan, C, Li, R, Huang, M.D. | | Deposit date: | 2018-11-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structural determination of group A Streptococcal surface dehydrogenase and characterization of its interaction with urokinase-type plasminogen activator receptor.
Biochem.Biophys.Res.Commun., 510, 2019
|
|
6K0L
 
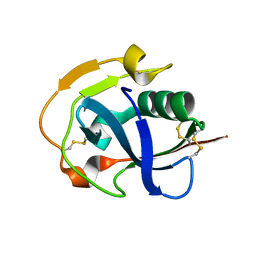 | | The crystal structure of simian CD163 SRCR5 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
6K0O
 
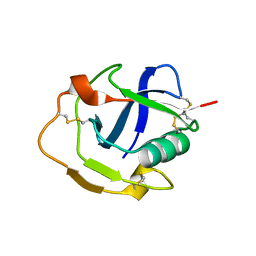 | | The crystal structure of human CD163-like homolog SRCR8 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M160 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
7WC0
 
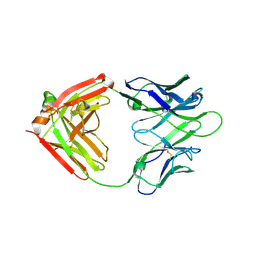 | |
7WCD
 
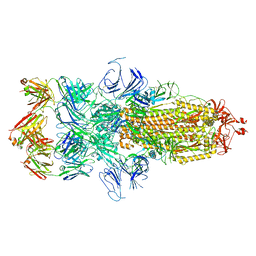 | | Cryo EM structure of SARS-CoV-2 spike in complex with TAU-2212 mAbs in conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Xiang, Y, Ma, B, Li, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
7C9Q
 
 | | Crystal structure of Human liver fructose-1,6-bisphoaphatase complex with Mg2+ and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION | | Authors: | Huang, Y, Li, R, Wan, J. | | Deposit date: | 2020-06-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Crystal structure of Human liver fructose-1,6-bisphoaphatase complex with Mg2+ and AMP
To Be Published
|
|
