1DMK
 
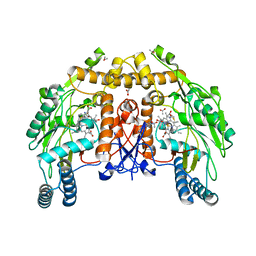 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 4-AMINO-6-PHENYL-TETRAHYDROPTERIDINE | | Descriptor: | 2,4-DIAMINO-6-PHENYL-5,6,7,8,-TETRAHYDROPTERIDINE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
1DMI
 
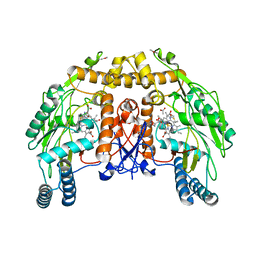 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 6S-H4B | | Descriptor: | 6S-5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kotsonis, P, Pfleiderer, W, Schmidt, H.H.H.W, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bovine Endothelial Nitric Oxide Synthase Heme Domain Complexed with the Pterin Analogues
To be Published
|
|
1RS7
 
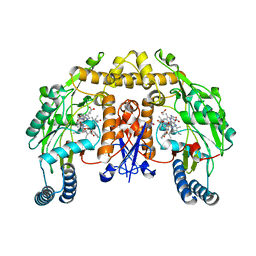 | | Rat neuronal NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
3ER8
 
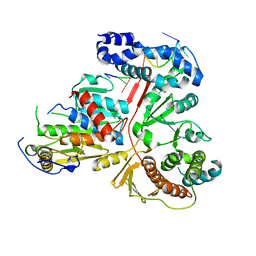 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with two fragments of RNA | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit, RNA/DNA chimera 5'-D(CP*CP*)R(UP*UP*)D(C)-3', ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
1Q2O
 
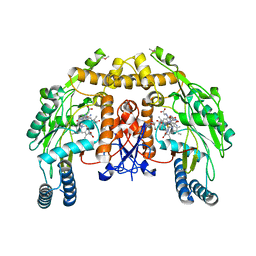 | | Bovine endothelial nitric oxide synthase N368D mutant heme domain dimer with L-N(omega)-nitroarginine-2,4-L-diaminobutyramide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-07-25 | | Release date: | 2004-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6LKW
 
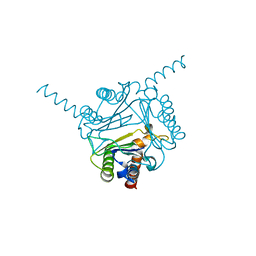 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
1SN4
 
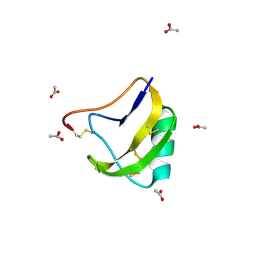 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
4R0X
 
 | | Allosteric coupling of conformational transitions in the FK1 domain of FKBP51 near the site of steroid receptor interaction | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | LeMaster, D.M, Mustafi, S.M, Brecher, M, Zhang, J, Heroux, A, Li, H.M, Hernandez, G. | | Deposit date: | 2014-08-02 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coupling of Conformational Transitions in the N-terminal Domain of the 51-kDa FK506-binding Protein (FKBP51) Near Its Site of Interaction with the Steroid Receptor Proteins.
J.Biol.Chem., 290, 2015
|
|
5GIT
 
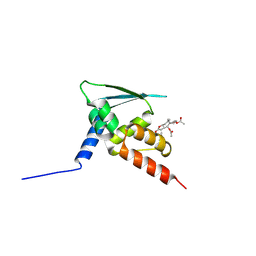 | | BTB domain of KEAP1 in complex with XX3 | | Descriptor: | Kelch-like ECH-associated protein 1, [(3aS,5R,5aS,6S,8S,8aS,9S,9aR)-9-acetyloxy-8-hydroxy-5,8a-dimethyl-1-methylidene-2-oxo-4,5,5a,6,7,8,9,9a-octahydro-3aH-azuleno[6,5-b]furan-6-yl] acetate | | Authors: | Zhu, L.L, Li, H.L, Wu, F.S, Xiong, R. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Britanin Ameliorates Cerebral Ischemia-Reperfusion Injury by Inducing the Nrf2 Protective Pathway.
Antioxid. Redox Signal., 27, 2017
|
|
6WNK
 
 | | Macrocyclic peptides TDI5575 that selectively inhibit the Mycobacterium tuberculosis proteasome | | Descriptor: | (12S,15S)-N-[(2-fluorophenyl)methyl]-10,13-dioxo-12-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}-2-oxa-11,14-diazatricyclo[15.2.2.1~3,7~]docosa-1(19),3(22),4,6,17,20-hexaene-15-carboxamide, CITRIC ACID, DIMETHYLFORMAMIDE, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Macrocyclic Peptides that Selectively Inhibit the Mycobacterium tuberculosis Proteasome.
J.Med.Chem., 64, 2021
|
|
8DWJ
 
 | |
6M2B
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with S416 | | Descriptor: | 2-[(E)-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]-methyl-hydrazinylidene]methyl]benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2.
Protein Cell, 11, 2020
|
|
6XN4
 
 | | Structure of the Lactococcus lactis Csm CTR_3:2 CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XN5
 
 | | Structure of the Lactococcus lactis Csm Apo- CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm3, CRISPR-associated protein Csm4, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XN3
 
 | | Structure of the Lactococcus lactis Csm CTR_4:3 CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XN7
 
 | | Structure of the Lactococcus lactis Csm NTR CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
2RFK
 
 | | Substrate RNA Positioning in the Archaeal H/ACA Ribonucleoprotein Complex | | Descriptor: | Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, Small nucleolar rnp similar to gar1, ... | | Authors: | Liang, B, Xue, S, Terns, R.M, Terns, M.P, Li, H, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-09-30 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1NDM
 
 | |
8FOJ
 
 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the post RNA handoff state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOC
 
 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase in Apo state conformation I | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
7BU9
 
 | | Crystal Structure of Spindlin1-H3(K4me3-K9me2) complex | | Descriptor: | H3(K4me3-K9me2) peptide, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Molecular basis for histone H3 "K4me3-K9me3/2" methylation pattern readout by Spindlin1.
J.Biol.Chem., 295, 2020
|
|
8FOH
 
 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the RNA synthesis state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
7BQZ
 
 | |
8FOE
 
 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex bound to a template DNA | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOK
 
 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the DNA elongation state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
