7EVR
 
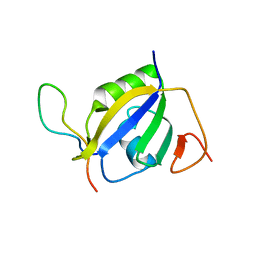 | | Crystal structure of hnRNP L RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L, SHI domain from Histone-lysine N-methyltransferase SETD2 | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7EVS
 
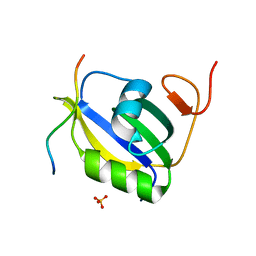 | | Crystal structure of hnRNP LL RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L-like, SHI domain from Histone-lysine N-methyltransferase SETD2, SULFATE ION | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
6KI6
 
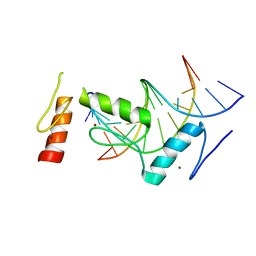 | | Crystal structure of BCL11A in complex with gamma-globin -115 HPFH region | | Descriptor: | B-cell lymphoma/leukemia 11A, DNA (5'-D(*AP*TP*AP*TP*TP*GP*GP*TP*CP*AP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*GP*AP*CP*CP*AP*AP*TP*A)-3'), ... | | Authors: | Li, F.D, Yang, Y, Shi, Y.Y. | | Deposit date: | 2019-07-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the recognition of gamma-globin gene promoter by BCL11A.
Cell Res., 29, 2019
|
|
8VA1
 
 | |
8V33
 
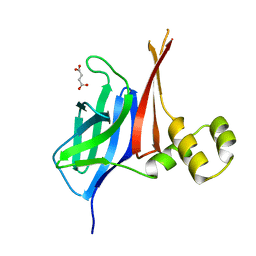 | |
8WU6
 
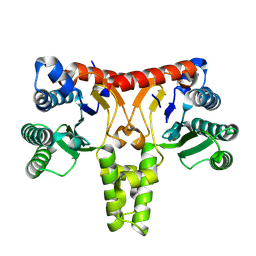 | | Structure of a Nerylneryl Diphosphate Synthase from Solanum lycopersicum | | Descriptor: | Nerylneryl diphosphate synthase CPT2, chloroplastic | | Authors: | Li, F.R, Wang, Q.L, Pan, X.M, Dong, L.B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery, Structure, and Engineering of a cis-Geranylfarnesyl Diphosphate Synthase.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8WU7
 
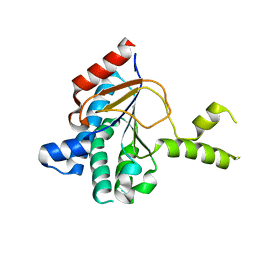 | | Structure of a cis-Geranylfarnesyl Diphosphate Synthase from Streptomyces clavuligerus | | Descriptor: | Isoprenyl transferase | | Authors: | Li, F.R, Wang, Q.L, Pan, X.M, Dong, L.B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery, Structure, and Engineering of a cis-Geranylfarnesyl Diphosphate Synthase.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4GNG
 
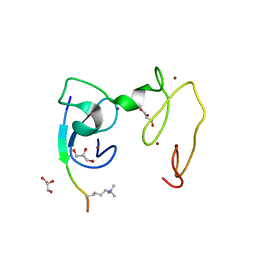 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GND
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains | | Descriptor: | Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4H14
 
 | | Crystal Structure of Bovine Coronavirus Spike Protein Lectin Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Li, F. | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of bovine coronavirus spike protein lectin domain.
J.Biol.Chem., 287, 2012
|
|
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNE
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
8HR5
 
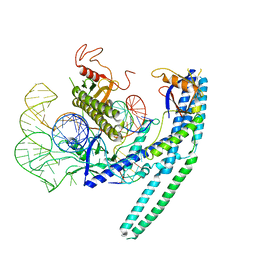 | | Cryo-EM structure of the CnCas12f1-sgRNA-DNA complex | | Descriptor: | DNA (28-MER), DNA (5'-D(*TP*AP*AP*CP*CP*TP*AP*AP*TP*AP*GP*AP*TP*GP*TP*GP*AP*A)-3'), Transposase, ... | | Authors: | Li, F, Ji, Q. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure and engineering of miniature Clostridium novyi CRISPR-Cas12f1 with rare C-rich PAM specificity
To Be Published
|
|
5YB5
 
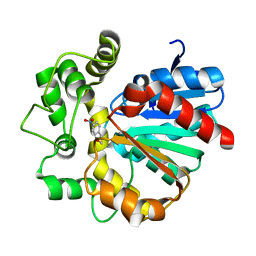 | | The complex crystal structure of VrEH2 mutant M263N with SNO | | Descriptor: | (S)-PARA-NITROSTYRENE OXIDE, Epoxide hydrolase | | Authors: | Li, F.L, Chen, F.F, Chen, Q, Kong, X.D, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-09-03 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
5YJ3
 
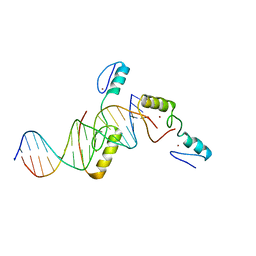 | | Crystal structure of TZAP and telomeric DNA complex | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), Telomere zinc finger-associated protein, ... | | Authors: | Li, F, Zhao, Y. | | Deposit date: | 2017-10-07 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | The 11th C2H2 zinc finger and an adjacent C-terminal arm are responsible for TZAP recognition of telomeric DNA.
Cell Res., 28, 2018
|
|
5Y6Y
 
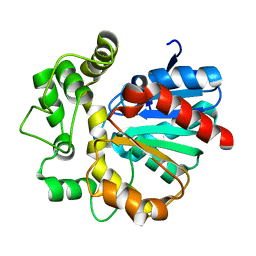 | | The crystal structure of VrEH2 mutant M263N | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Yu, H.L, Chen, Q, Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-08-15 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
7UKJ
 
 | |
6AKJ
 
 | | The crystal structure of EMC complex | | Descriptor: | Enhancer of rudimentary homolog,YTH domain-containing protein mmi1 fusion protein, SULFATE ION | | Authors: | Li, F. | | Deposit date: | 2018-09-01 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved dimer interface connects ERH and YTH family proteins to promote gene silencing.
Nat Commun, 10, 2019
|
|
5XM6
 
 | | the overall structure of VrEH2 | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Kong, X.D, Yu, H.L, Shang, Y.P, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
7CG2
 
 | | Vigna radiata Epoxide hydrolase mutant | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Reprogramming Epoxide Hydrolase to Improve Enantioconvergence in Hydrolysis of Styrene Oxide Scaffolds
Adv.Synth.Catal., 362, 2021
|
|
7CG6
 
 | | Vigna radiata Epoxide hydrolase mutant M263Q | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reprogramming Epoxide Hydrolase to Improve Enantioconvergence in Hydrolysis of Styrene Oxide Scaffolds
Adv.Synth.Catal., 362, 2021
|
|
7DBE
 
 | | Structure of a novel transaminase | | Descriptor: | 1,2-ETHANEDIOL, PYRIDOXAL-5'-PHOSPHATE, branched-chain amino acid aminotransferase | | Authors: | Li, F.L, Yu, H.M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of an (R)-Selective Transaminase in the Asymmetric Synthesis of Chiral Hydroxy Amines.
Adv.Synth.Catal., 2021
|
|
7CNC
 
 | | cystal structure of human ERH in complex with DGCR8 | | Descriptor: | Enhancer of rudimentary homolog, Microprocessor complex subunit DGCR8 | | Authors: | Li, F, Shen, S. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8.
Nucleic Acids Res., 48, 2020
|
|
3KBH
 
 | | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Wu, K, Li, W, Peng, G, Li, F. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5JCP
 
 | | RhoGAP domain of ARAP3 in complex with RhoA in the transition state | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3,Linker,Transforming protein RhoA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bao, H, Li, F, Wang, C, Wang, N, Jiang, Y, Tang, Y, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
