7WDW
 
 | | DsyB in complex with SAH and MTHB | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-methylsulfanyl-2-oxidanyl-butanoic acid, DSYB, ... | | Authors: | Li, C.Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Mechanistic insights into the key marine dimethylsulfoniopropionate synthesis enzyme DsyB/DSYB.
Mlife, 2022
|
|
4QFK
 
 | |
4QFL
 
 | |
4QFO
 
 | |
1BXZ
 
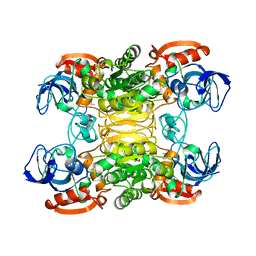 | | CRYSTAL STRUCTURE OF A THERMOPHILIC ALCOHOL DEHYDROGENASE SUBSTRATE COMPLEX FROM THERMOANAEROBACTER BROCKII | | Descriptor: | 2-BUTANOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, C, Heatwole, J, Soelaiman, S, Shoham, M. | | Deposit date: | 1998-10-09 | | Release date: | 2000-02-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of a thermophilic alcohol dehydrogenase substrate complex suggests determinants of substrate specificity and thermostability.
Proteins, 37, 1999
|
|
4QFP
 
 | |
6KY8
 
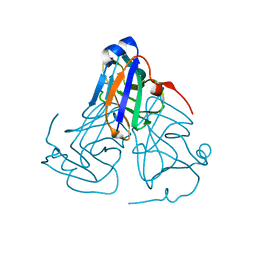 | | Crystal structure of ASFV dUTPase | | Descriptor: | E165R | | Authors: | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
4QFN
 
 | |
3EIP
 
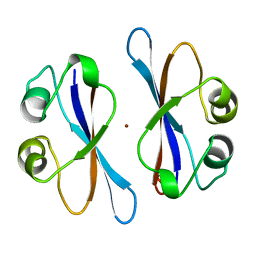 | | CRYSTAL STRUCTURE OF COLICIN E3 IMMUNITY PROTEIN: AN INHIBITOR TO A RIBOSOME-INACTIVATING RNASE | | Descriptor: | PROTEIN (COLICIN E3 IMMUNITY PROTEIN), ZINC ION | | Authors: | Li, C, Zhao, D, Djebli, A, Shoham, M. | | Deposit date: | 1999-03-29 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase.
Structure Fold.Des., 7, 1999
|
|
7D4N
 
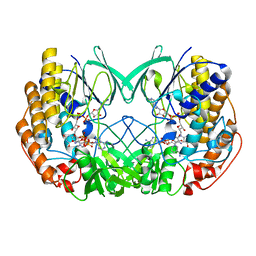 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 20 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
7D4M
 
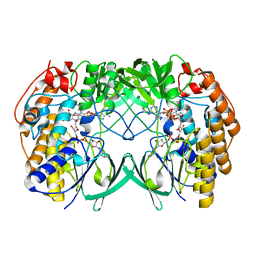 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 5 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
7D4K
 
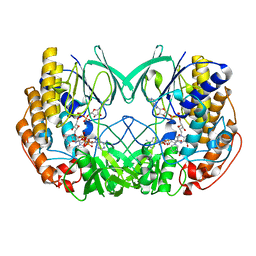 | | Crystal structure of Tmm from Pelagibacter sp. strain HTCC7211 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
4KJI
 
 | |
8OS5
 
 | | Crystal structure of the Factor XII heavy chain reveals an interlocking dimer with a FnII to kringle domain interaction | | Descriptor: | Coagulation factor XII-Mie | | Authors: | Li, C, Saleem, M, Kaira, B.G, Brown, A, Wilson, C, Philippou, H, Emsley, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Factor XII and kininogen asymmetric assembly with gC1qR/C1QBP/P32 is governed by allostery.
Blood, 136, 2020
|
|
6JZP
 
 | |
6K9X
 
 | | Crystal Structure Analysis of Protein | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-18 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism
Acta Crystallographica Section D-Biological Crystallography, D70, 2014
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5WYB
 
 | | Structure of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
5WYD
 
 | | Structural of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
6UGH
 
 | |
5WZE
 
 | | The structure of Pseudomonas aeruginosa aminopeptidase PepP | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aminopeptidase P, ... | | Authors: | Bao, R, Peng, C.T, Liu, L, He, L.H, Li, C.C, Li, T, Shen, Y.L, Zhu, Y.B, Song, Y.J. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa.
Front Microbiol, 8, 2017
|
|
1QX3
 
 | | Conformational restrictions in the active site of unliganded human caspase-3 | | Descriptor: | Apopain | | Authors: | Ni, C.-Z, Li, C, Wu, J.C, Spada, A.P, Ely, K.R. | | Deposit date: | 2003-09-04 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational restrictions in the active site of unliganded human caspase-3
J.MOL.RECOG., 16, 2003
|
|
4N6X
 
 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1/Chemokine Receptor CXCR2 fusion protein | | Authors: | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.051 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
2A25
 
 | | Crystal structure of Siah1 SBD bound to the peptide EKPAAVVAPITTG from SIP | | Descriptor: | Calcyclin-binding protein peptide, Ubiquitin ligase SIAH1, ZINC ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
1Q6T
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 6-[4-((2S)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-[(1S)-1-METHOXY-3-METHYLBUTYL]QUINOLIN-8-YLPHOSPHONIC ACID, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
