4J34
 
 | | Crystal Structure of kynurenine 3-monooxygenase - truncated at position 394 plus HIS tag cleaved. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J33
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-394) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
5M0O
 
 | | Crystal structure of cytochrome P450 OleT H85Q in complex with arachidonic acid | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
5M0P
 
 | | Crystal structure of cytochrome P450 OleT F79A in complex with arachidonic acid | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, Terminal olefin-forming fatty acid decarboxylase, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
1D7O
 
 | | CRYSTAL STRUCTURE OF BRASSICA NAPUS ENOYL ACYL CARRIER PROTEIN REDUCTASE COMPLEXED WITH NAD AND TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Roujeinikova, A, Levy, C, Rowsell, S, Sedelnikova, S, Baker, P.J, Minshull, C.A, Mistry, A, Colls, J.G, Camble, R, Stuitje, A.R, Slabas, A.R, Rafferty, J.B, Pauptit, R.A, Viner, R, Rice, D.W. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of triclosan bound to enoyl reductase.
J.Mol.Biol., 294, 1999
|
|
5MHK
 
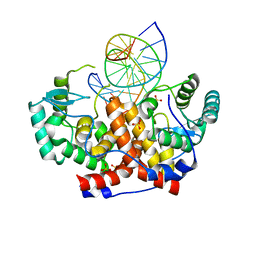 | | ICP4 DNA-binding domain in complex with 19mer DNA duplex from its own promoter | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*TP*CP*CP*GP*TP*GP*TP*GP*GP*AP*CP*GP*AP*TP*CP*GP*G)-3'), ICP4 DNA BINDING DOMAIN, ... | | Authors: | Tunnicliffe, R.B, Lockhart-Cairns, M.P, Levy, C, Mould, P, Jowitt, T.A, Sito, H, Baldock, C, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res., 45, 2017
|
|
5MHJ
 
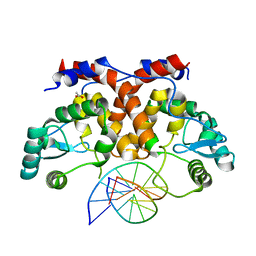 | | ICP4 DNA-binding domain, lacking intrinsically disordered region, in complex with 12mer DNA duplex from its own promoter | | Descriptor: | ACETIC ACID, CHLORIDE ION, DNA (5'-D(P*CP*GP*AP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Tunnicliffe, R.B, Lockhart-Cairns, M.P, Levy, C, Mould, P, Jowitt, T.A, Sito, H, Baldock, C, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res., 45, 2017
|
|
3GSI
 
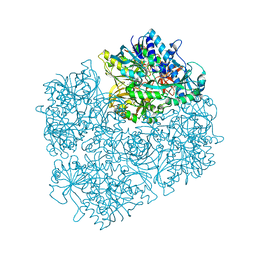 | | Crystal structure of D552A dimethylglycine oxidase mutant of Arthrobacter globiformis in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tralau, T, Lafite, P, Levy, C, Combe, J.P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An internal reaction chamber in dimethylglycine oxidase provides efficient protection from exposure to toxic formaldehyde.
J.Biol.Chem., 284, 2009
|
|
3IA5
 
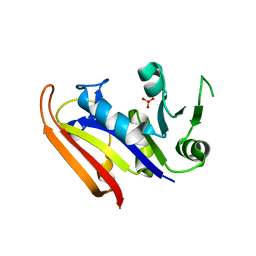 | | Moritella profunda dihydrofolate reductase (DHFR) | | Descriptor: | Dihydrofolate reductase, PHOSPHATE ION | | Authors: | Hay, S, Evans, R.M, Levy, C, Wang, X, Loveridge, E.J, Leys, D, Allemann, R.K, Scrutton, N.S. | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Are the Catalytic Properties of Enzymes from Piezophilic Organisms Pressure Adapted?
Chembiochem, 10, 2009
|
|
3IVY
 
 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125, p212121 crystal form | | Descriptor: | Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
3IW2
 
 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125 in complex with econazole | | Descriptor: | 4-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
3IP8
 
 | | Crystal structure of arylmalonate decarboxylase (AMDase) from Bordatella bronchiseptic in complex with benzylphosphonate | | Descriptor: | Arylmalonate decarboxylase, benzylphosphonic acid | | Authors: | Okrasa, K, Levy, C, Leys, D, Micklefield, J. | | Deposit date: | 2009-08-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure-Guided Directed Evolution of Alkenyl and Arylmalonate Decarboxylases.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IW0
 
 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125, C2221 crystal form | | Descriptor: | Cytochrome P450 CYP125, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
3IW1
 
 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125 in complex with androstenedione | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
3JSX
 
 | | X-ray Crystal structure of NAD(P)H: Quinone Oxidoreductase-1 (NQO1) bound to the coumarin-based inhibitor AS1 | | Descriptor: | 4-hydroxy-6,7-dimethyl-3-(naphthalen-1-ylmethyl)-2H-chromen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Dunstan, M.S, Levy, C, Leys, D. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1).
J.Med.Chem., 52, 2009
|
|
6Y1T
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a Trp51 to S-Trp51 modification | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
6Y2Y
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with Trp51 to S-Trp51 and Trp191Phe modifications | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
7ZP7
 
 | | Crystal structure of evolved photoenzyme EnT1.3 (truncated) with bound product | | Descriptor: | (1~{R},10~{R},12~{S})-15-oxa-8-azatetracyclo[8.5.0.0^{1,12}.0^{2,7}]pentadeca-2(7),3,5-trien-9-one, 1,2-ETHANEDIOL, EnT1.3 C | | Authors: | Hardy, F.J, Levy, C. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A designed photoenzyme for enantioselective [2+2] cycloadditions.
Nature, 611, 2022
|
|
7ZP6
 
 | |
7ZP5
 
 | |
5M0N
 
 | | Crystal structure of cytochrome P450 OleT in complex with formate | | Descriptor: | FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
7QQ7
 
 | |
8C3W
 
 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
6FAD
 
 | | SR protein kinase 1 (SRPK1) in complex with the RGG-box of HSV1 ICP27 | | Descriptor: | PHOSPHATE ION, SRSF protein kinase 1, mRNA export factor | | Authors: | Tunnicliffe, R.B, Levy, C, Mould, A.P, Mckenzie, E.A, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2017-12-15 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Molecular Mechanism of SR Protein Kinase 1 Inhibition by the Herpes Virus Protein ICP27.
Mbio, 10, 2019
|
|
6H08
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a His175Me-His proximal ligand substitution | | Descriptor: | COBALT (II) ION, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2018-07-06 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rewiring the "Push-Pull" Catalytic Machinery of a Heme Enzyme Using an Expanded Genetic Code.
Acs Catalysis, 10, 2020
|
|
