7DA5
 
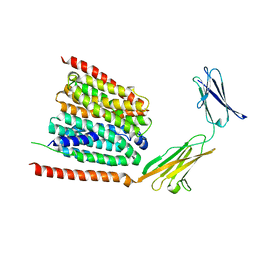 | | Cryo-EM structure of the human MCT1 D309N mutant in complex with Basigin-2 in the inward-open conformation. | | Descriptor: | Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
7DD3
 
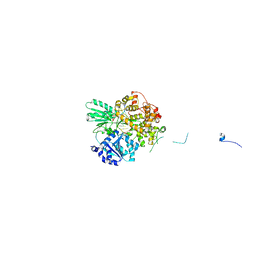 | | Cryo-EM structure of the pre-mRNA-loaded DEAH-box ATPase/helicase Prp2 in complex with Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2, pre-mRNA | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DCP
 
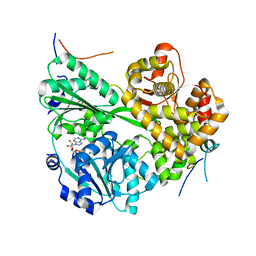 | | cryo-EM structure of the DEAH-box helicase Prp2 and coactivator Spp2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PRP2 isoform 1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DCO
 
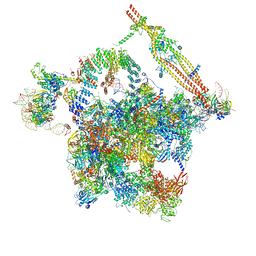 | | Cryo-EM structure of the activated spliceosome (Bact complex) at an atomic resolution of 2.5 angstrom | | Descriptor: | BJ4_G0014900.mRNA.1.CDS.1, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0037700.mRNA.1.CDS.1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DVQ
 
 | | Cryo-EM Structure of the Activated Human Minor Spliceosome (minor Bact Complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'-O-[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(methoxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Armadillo repeat-containing protein 7, ... | | Authors: | Bai, R, Wan, R, Wang, L, Xu, K, Zhang, Q, Lei, J, Shi, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the activated human minor spliceosome.
Science, 371, 2021
|
|
7FAY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with (R)-1a | | Descriptor: | (2~{R})-~{N}-[(1~{R})-2-(~{tert}-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-~{N}-(4-~{tert}-butylphenyl)-2-oxidanyl-propanamide, 3C-like proteinase | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
7FAZ
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Y180 | | Descriptor: | (2~{R})-~{N}-dibenzofuran-3-yl-~{N}-[(1~{R})-2-[[(1~{S})-1-(4-fluorophenyl)ethyl]amino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]-2-oxidanyl-propanamide, 3C-like proteinase, SODIUM ION | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
7WZO
 
 | |
7WSW
 
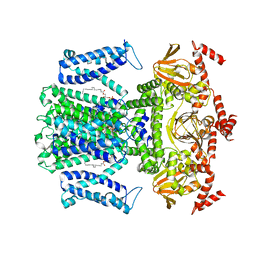 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7XXK
 
 | |
7YBG
 
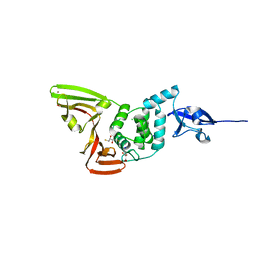 | |
7XUF
 
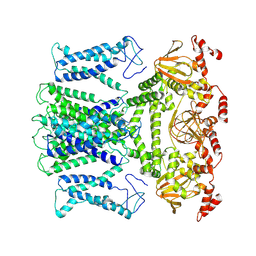 | | Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana | | Descriptor: | POTASSIUM ION, Potassium channel AKT1, Potassium channel KAT3 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Yang, F, Zhang, Y.M, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7Y7Y
 
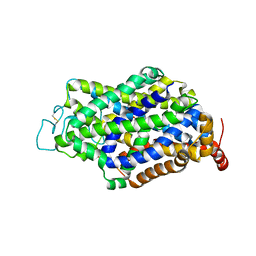 | | Cryo-EM structure of human GABA transporter GAT1 bound with nipecotic acid in NaCl solution in an inward-occluded state at 2.4 angstrom | | Descriptor: | (3R)-piperidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y7V
 
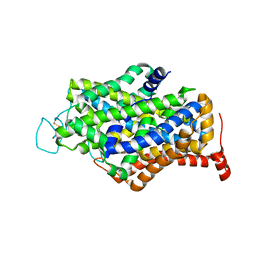 | | Cryo-EM structure of human apo GABA transporter GAT1 in an inward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Sodium- and chloride-dependent GABA transporter 1 | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y7W
 
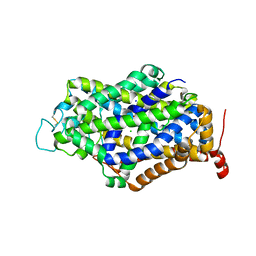 | | Cryo-EM structure of human GABA transporter GAT1 bound with GABA in NaCl solution in an inward-occluded state at 2.4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y7Z
 
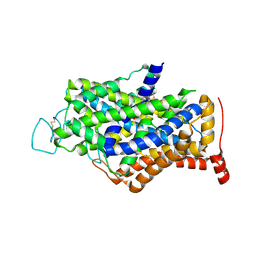 | | Cryo-EM structure of human GABA transporter GAT1 bound with tiagabine in NaCl solution in an inward-open state at 3.2 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Sodium- and chloride-dependent GABA transporter 1, ... | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y5X
 
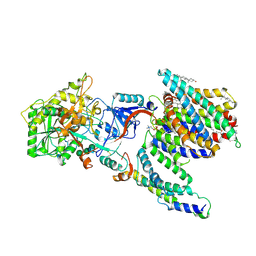 | | CryoEM structure of PS2-containing gamma-secretase treated with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5T
 
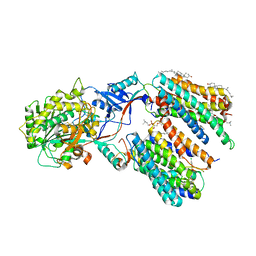 | | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5Z
 
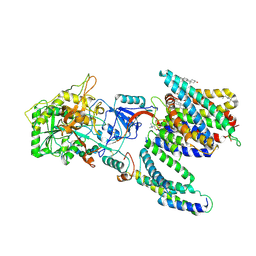 | | CryoEM structure of human PS2-containing gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7WX2
 
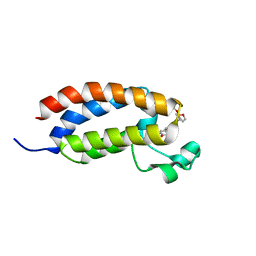 | | CBP-BrD complexed with NEO2734 | | Descriptor: | 1,3-dimethyl-5-[2-(oxan-4-yl)-3-[2-(trifluoromethyloxy)ethyl]benzimidazol-5-yl]pyridin-2-one, CREB-binding protein | | Authors: | Zeng, L, Lei, J.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Targeting CDCP1 gene transcription coactivated by BRD4 and CBP/p300 in castration-resistant prostate cancer.
Oncogene, 41, 2022
|
|
7WWZ
 
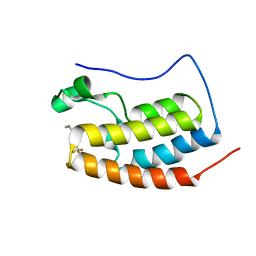 | | BRD4-BD1 complexed with NEO2734 | | Descriptor: | 1,3-dimethyl-5-[2-(oxan-4-yl)-3-[2-(trifluoromethyloxy)ethyl]benzimidazol-5-yl]pyridin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Zeng, L, Lei, J.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Targeting CDCP1 gene transcription coactivated by BRD4 and CBP/p300 in castration-resistant prostate cancer.
Oncogene, 41, 2022
|
|
2V3W
 
 | | Crystal structure of the benzoylformate decarboxylase variant L461A from Pseudomonas putida | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Gocke, D, Walter, L, Gauchenova, K, Kolter, G, Knoll, M, Berthold, C.L, Schneider, G, Pleiss, J, Mueller, M, Pohl, M. | | Deposit date: | 2007-06-25 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Protein Design of Thdp-Dependent Enzymes-Engineering Stereoselectivity.
Chembiochem, 9, 2008
|
|
6FHE
 
 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Synthetic construct | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
6FHF
 
 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Design, SODIUM ION | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
8A9Q
 
 | | Computational design of stable mammalian serum albumins for bacterial expression | | Descriptor: | Albumin, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Khersonsky, O, Dym, O, Fleishman, J.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stable Mammalian Serum Albumins Designed for Bacterial Expression.
J.Mol.Biol., 435, 2023
|
|
