1CQ0
 
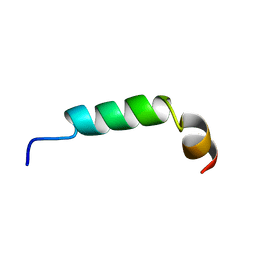 | | SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B'SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B ' | | Descriptor: | PROTEIN (NEW HYPOTHALAMIC NEUROPEPTIDE/OREXIN-B28) | | Authors: | Lee, K.-H, Bang, E.J, Chae, K.-J, Lee, D.W, Lee, W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-01-10 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a new hypothalamic neuropeptide, human hypocretin-2/orexin-B.
Eur.J.Biochem., 266, 1999
|
|
1IQO
 
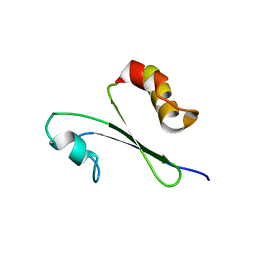 | | Solution structure of MTH1880 from methanobacterium thermoautotrophicum | | Descriptor: | HYPOTHETICAL PROTEIN MTH1880 | | Authors: | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2001-07-23 | | Release date: | 2002-07-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
6XE4
 
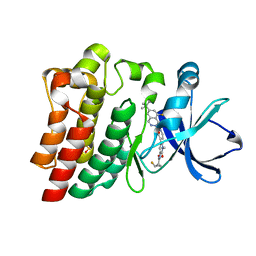 | | BTK Fluorocyclopropyl amide inhibitor, Compound 25 | | Descriptor: | (1S,2S)-N-[2'-(6-tert-butyl-8-fluoro-1-oxophthalazin-2(1H)-yl)-3'-(hydroxymethyl)-1-methyl-6-oxo[1,6-dihydro[3,4'-bipyridine]]-5-yl]-2-fluorocyclopropane-1-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Kiefer, J.R, Crawford, J.J, Lee, W, Eigenbrot, C, Yu, C. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereochemical Differences in Fluorocyclopropyl Amides Enable Tuning of Btk Inhibition and Off-Target Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
8HU2
 
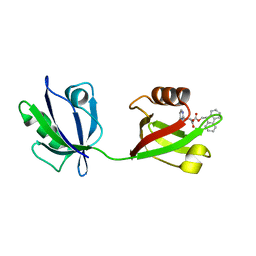 | | Rattus Syntenin-1 PDZ domain with inhibitor | | Descriptor: | (2~{S})-2-(9~{H}-fluoren-9-ylmethoxycarbonylamino)-3-(4-oxidanylidene-5~{H}-pyrimidin-2-yl)propanoic acid, Syntenin-1 | | Authors: | Heo, Y, Lee, J, Yun, J.H, Lee, W. | | Deposit date: | 2022-12-22 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of STNPDZ with inhibitor at 1.60 Angstroms resolution.
To Be Published
|
|
1IQS
 
 | | Minimized average structure of MTH1880 from Methanobacterium Thermoautotrophicum | | Descriptor: | MTH1880 | | Authors: | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2001-07-29 | | Release date: | 2002-07-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
6CGH
 
 | | Solution structure of the four-helix bundle region of human J-protein Zuotin, a component of ribosome-associated complex (RAC) | | Descriptor: | DnaJ homolog subfamily C member 2 | | Authors: | Shrestha, O.K, Lee, W, Tonelli, M, Cornilescu, G, Markley, J.L, Ciesielski, S.J, Craig, E.A. | | Deposit date: | 2018-02-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and evolution of the 4-helix bundle domain of Zuotin, a J-domain protein co-chaperone of Hsp70.
Plos One, 14, 2019
|
|
1RJA
 
 | |
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8HNJ
 
 | | Domain-stabilized glutamine-binding protein | | Descriptor: | GLUTAMINE, Glutamine ABC transporter, periplasmic glutamine-binding protein GlnH | | Authors: | Choi, S.H, Park, J.H, Seo, M.H, Park, K.W, Lee, W.K. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Domain-wise dissection of thermal stability enhancement in multidomain proteins.
Int.J.Biol.Macromol., 237, 2023
|
|
4ZG0
 
 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
6L7Z
 
 | | Solution NMR structure of the N-terminal immunoglobulin variable domain of BTNL2 | | Descriptor: | Butyrophilin-like protein 2 | | Authors: | Basak, A.J, Lee, W, Samanta, D, De, S. | | Deposit date: | 2019-11-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into N-terminal IgV Domain of BTNL2, a T Cell Inhibitory Molecule, Suggests a Non-canonical Binding Interface for Its Putative Receptors.
J.Mol.Biol., 432, 2020
|
|
6LVU
 
 | |
7X0D
 
 | | Crystal structure of phospholipase A1, CaPLA1 | | Descriptor: | Phospholipase A1, SULFATE ION | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39725161 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
7X0C
 
 | | Crystal structure of phospholipase A1, AtDSEL | | Descriptor: | Phospholipase A1-IIgamma | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79909515 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
3K9P
 
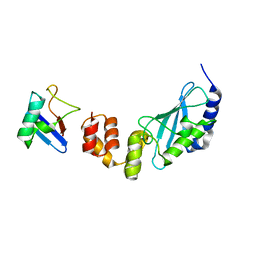 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
1F5V
 
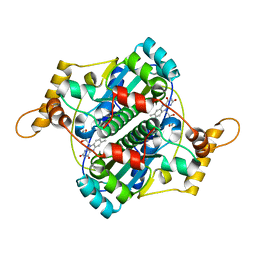 | | STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF A FLAVOPROTEIN FROM ESCHERICHIA COLI THAT REDUCES NITROCOMPOUNDS. ALTERATION OF PYRIDINE NUCLEOTIDE BINDING BY A SINGLE AMINO ACID SUBSTITUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NADPH NITROREDUCTASE | | Authors: | Kobori, T, Sasaki, H, Lee, W.C, Zenno, S, Saigo, K, Murphy, M.E.P, Tanokura, M. | | Deposit date: | 2000-06-17 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.
J.Biol.Chem., 276, 2001
|
|
6OCV
 
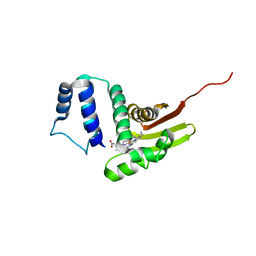 | |
2NLU
 
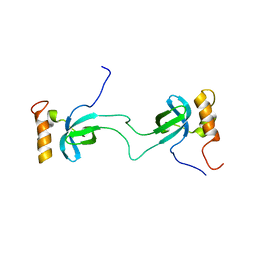 | |
2GKI
 
 | | Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity | | Descriptor: | nuclease | | Authors: | Kim, Y.R, Kim, J.S, Lee, S.H, Lee, W.R, Sohn, J.N, Chung, Y.C, Shim, H.K, Lee, S.C, Kwon, M.H, Kim, Y.S. | | Deposit date: | 2006-04-02 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity.
J.Biol.Chem., 281, 2006
|
|
2KCC
 
 | | Solution Structure of biotinoyl domain from human acetyl-CoA carboxylase 2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Lee, C, Cheong, H, Ryu, K, Lee, J, Lee, W, Jeon, Y, Cheong, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins, 72, 2008
|
|
1WPI
 
 | | Solution NMR Structure of Protein YKR049C from Saccharomyces cerevisiae. Ontario Centre for Structural Proteomics target YST0250_1_133; Northeast Structural Genomics Consortium YTYst250 | | Descriptor: | Hypothetical 15.6 kDa protein in NAP1-TRK2 intergenic region | | Authors: | Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-03 | | Release date: | 2005-09-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of YKR049C, a putative redox protein from Saccharomyces cerevisiae
J.Biochem.Mol.Biol., 38, 2005
|
|
7LHQ
 
 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | Descriptor: | Non-structural protein 7 | | Authors: | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
6DTX
 
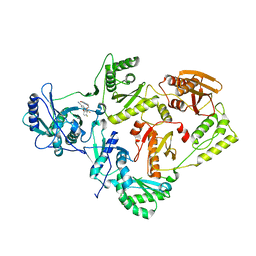 | | Wildtype HIV-1 Reverse Transcriptase in complex with JLJ 578 | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, GLYCEROL, Reverse transcriptase/ribonuclease H, ... | | Authors: | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.327 Å) | | Cite: | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6DTW
 
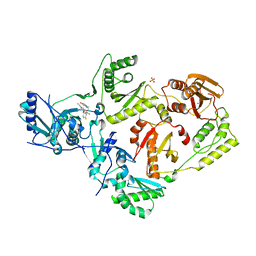 | | HIV-1 Reverse Transcriptase Y181C Mutant in complex with JLJ 578 | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5VSO
 
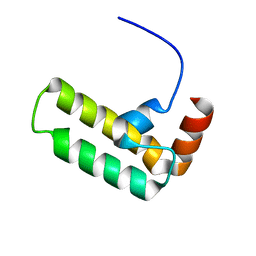 | | NMR structure of Ydj1 J-domain, a cytosolic Hsp40 from Saccharomyces cerevisiae | | Descriptor: | Yeast dnaJ protein 1 | | Authors: | Ciesielski, S.J, Tonelli, M, Lee, W, Cornilescu, G, Markley, J.L, Schilke, B.A, Ziegelhoffer, T, Craig, E.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Broadening the functionality of a J-protein/Hsp70 molecular chaperone system.
PLoS Genet., 13, 2017
|
|
