7U37
 
 | |
5DC7
 
 | | Crystal structure of D176A-Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | Fluor-de-Lys tetrapeptide assay substrate, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Lee, M.S, Christianson, D.W. | | Deposit date: | 2015-08-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | General Base-General Acid Catalysis in Human Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
5DC8
 
 | | Crystal structure of H142A-Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | Fluor-de-Lys tetrapeptide assay substrate, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Lee, M.S, Christianson, D.W. | | Deposit date: | 2015-08-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | General Base-General Acid Catalysis in Human Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
2Y2C
 
 | | crystal structure of AmpD Apoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y28
 
 | | crystal structure of Se-Met AmpD derivative | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2B
 
 | | crystal structure of AmpD in complex with reaction products | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
7EAR
 
 | | A positively charged mutant Cry3Aa endotoxin | | Descriptor: | Crystaline entomocidal protoxin | | Authors: | Yang, Z, Lee, M.M, Chan, M.K. | | Deposit date: | 2021-03-08 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficient intracellular delivery of p53 protein by engineered protein crystals restores tumor suppressing function in vivo.
Biomaterials, 271, 2021
|
|
6DLN
 
 | |
3LZC
 
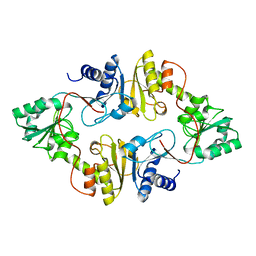 | | Crystal structure of Dph2 from Pyrococcus horikoshii | | Descriptor: | Dph2 | | Authors: | Zhang, Y, Zhu, X, Torelli, A.T, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
1XO3
 
 | | Solution Structure of Ubiquitin like protein from Mus Musculus | | Descriptor: | RIKEN cDNA 2900073H19 | | Authors: | Singh, S, Tonelli, M, Tyler, R.C, Bahrami, A, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-05 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the AAH26994.1 protein from Mus musculus, a putative eukaryotic Urm1.
Protein Sci., 14, 2005
|
|
7MG0
 
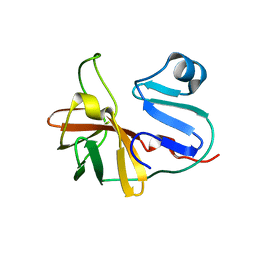 | |
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
5DC6
 
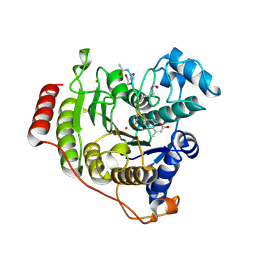 | | Crystal structure of D176N-Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | Fluor-de-Lys tetrapeptide assay substrate, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Lee, M.S, Christianson, D.W. | | Deposit date: | 2015-08-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | General Base-General Acid Catalysis in Human Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
3LZD
 
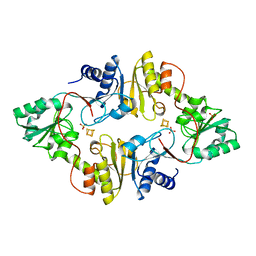 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
1XO8
 
 | | Solution structure of AT1g01470 from Arabidopsis Thaliana | | Descriptor: | At1g01470 | | Authors: | Singh, S, Cornilescu, C.C, Tyler, R.C, Cornilescu, G, Tonelli, M, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-06 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein
Protein Sci., 14, 2005
|
|
6AQF
 
 | | Crystal structure of A2AAR-BRIL in complex with the antagonist ZM241385 produced from Pichia pastoris | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Eddy, M.T, Lee, M.Y, Gao, Z, White, K, Didenko, T, Horst, R, Audet, M, Stanczak, P, McClary, K.M, Han, G.W, Jacobson, K.A, Stevens, R.C, Wuthrich, K. | | Deposit date: | 2017-08-19 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor.
Cell, 172, 2018
|
|
1Y4O
 
 | | Solution structure of a mouse cytoplasmic Roadblock/LC7 dynein light chain | | Descriptor: | Dynein light chain 2A, cytoplasmic | | Authors: | Song, J, Tyler, R.C, Lee, M.S, Tyler, E.M, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-01 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of isoform 1 of Roadblock/LC7, a light chain in the dynein complex.
J.Mol.Biol., 354, 2005
|
|
1T2V
 
 | | Structural basis of phospho-peptide recognition by the BRCT domain of BRCA1, structure with phosphopeptide | | Descriptor: | BRCTide-7PS, Breast cancer type 1 susceptibility protein | | Authors: | Williams, R.S, Lee, M.S, Hau, D.D, Glover, J.N.M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1
Nat.Struct.Mol.Biol., 11, 2004
|
|
3NIH
 
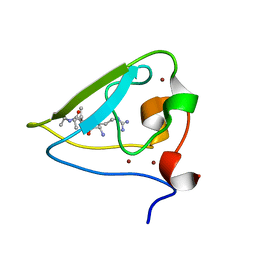 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIL
 
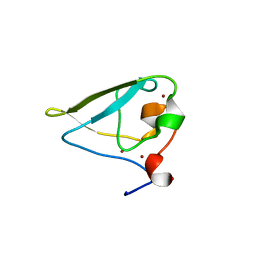 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
1CYZ
 
 | | NMR STRUCTURE OF THE GAACTGGTTC/TRI-IMIDAZOLE POLYAMIDE COMPLEX | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, 5'-D(*GP*AP*AP*CP*TP*GP*GP*TP*TP*C)-3' | | Authors: | Yang, X.-L, Hubbard IV, R.B, Lee, M, Tao, Z.-F, Sugiyama, H, Wang, A.H.-J. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Imidazole-imidazole pair as a minor groove recognition motif for T:G mismatched base pairs
Nucleic Acids Res., 27, 1999
|
|
1T2U
 
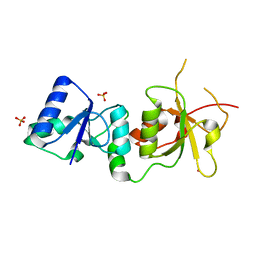 | | Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1: structure of BRCA1 missense variant V1809F | | Descriptor: | Breast cancer type 1 susceptibility protein, COBALT (II) ION, SULFATE ION | | Authors: | Williams, R.S, Lee, M.S, Duong, D.D, Glover, J.N.M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Phosphopeptide recognition by the BRCT domain of BRCA1
Nat.Struct.Mol.Biol., 11, 2004
|
|
6B73
 
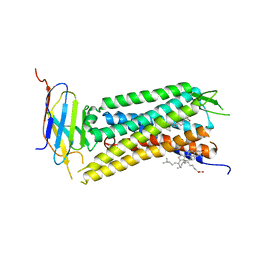 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
2WSY
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
5YKC
 
 | |
