8KA0
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | CALCIUM ION, Calmodulin-2, GLYCEROL, ... | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+)
To Be Published
|
|
8KA2
 
 | |
8K9Z
 
 | |
8KA1
 
 | |
6LVU
 
 | |
6KVX
 
 | | Crystal structure of the N-terminal domain single mutant (D119A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Endolysin,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Park, J, Lee, G, Yoon, S, Min, C.K, Kim, T.G, Yamamoto, T, Kim, D.H, Lee, K.W, Eom, S.H. | | Deposit date: | 2019-09-05 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the N-terminal domain single mutant (D119A) of the human mitochondrial calcium uniporter fused with T4 lysozyme
Sci Rep, 2020
|
|
4MRQ
 
 | | Crystal Structure of wild-type unphosphorylated PMM/PGM | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, ... | | Authors: | Lee, Y, Beamer, L. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Promotion of enzyme flexibility by dephosphorylation and coupling to the catalytic mechanism of a phosphohexomutase.
J.Biol.Chem., 289, 2014
|
|
5Y78
 
 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with inorganic phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PHOSPHATE ION, Putative hexose phosphate translocator | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
5Y79
 
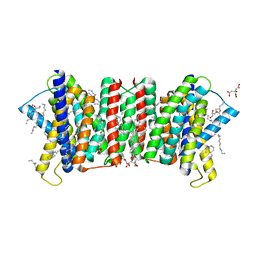 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with 3-phosphoglycerate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-PHOSPHOGLYCERIC ACID, CITRATE ANION, ... | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
4HQR
 
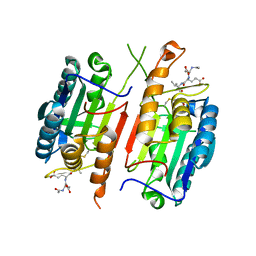 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Ac-Asp-Glu-Val-Asp-Aldehyde, Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-26 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HQ0
 
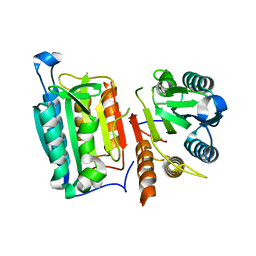 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-25 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6AGO
 
 | | Crystal structure of MRG15-ASH1L Histone methyltransferase complex | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, Mortality factor 4 like 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y, Song, J. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis of MRG15-Mediated Activation of the ASH1L Histone Methyltransferase by Releasing an Autoinhibitory Loop.
Structure, 27, 2019
|
|
6JMQ
 
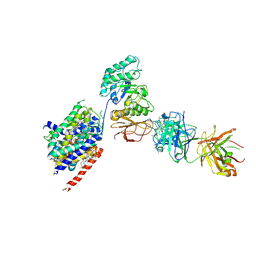 | | LAT1-CD98hc complex bound to MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Nakane, T, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JMR
 
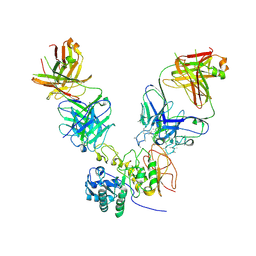 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2JTT
 
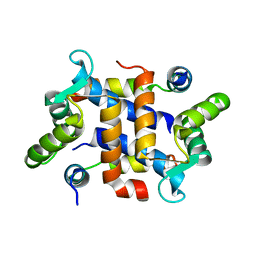 | |
6JG0
 
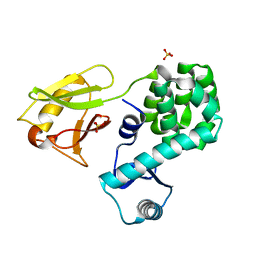 | | Crystal structure of the N-terminal domain single mutant (S92E) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Endolysin,Calcium uniporter protein, SULFATE ION | | Authors: | Lee, Y, Park, J, Min, C.K, Kang, J.Y, Kim, T.G, Yamamoto, T, Kim, D.H, Eom, S.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of calcium channel domain
To Be Published
|
|
6IF1
 
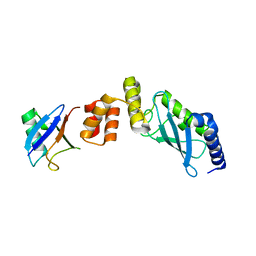 | | Crystal structure of Ube2K and K48-linked di-ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Lee, J.-G, Youn, H.-S, Lee, Y, An, J.Y, Park, K.R, Kang, J.Y, Lim, J.J, Eom, S.H. | | Deposit date: | 2018-09-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.466 Å) | | Cite: | Crystal structure of the Ube2K/E2-25K and K48-linked di-ubiquitin complex provides structural insight into the mechanism of K48-specific ubiquitin chain synthesis.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6GDG
 
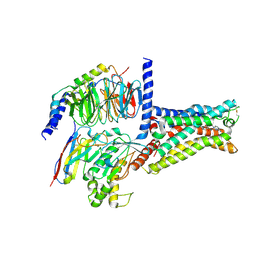 | | Cryo-EM structure of the adenosine A2A receptor bound to a miniGs heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Garcia-Nafria, J, Lee, Y. | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM structure of the adenosine A2Areceptor coupled to an engineered heterotrimeric G protein.
Elife, 7, 2018
|
|
7SIM
 
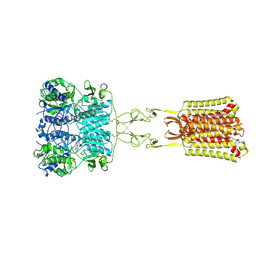 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIL
 
 | | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5EPP
 
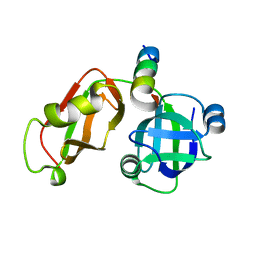 | | Structural Insights into the Interaction of p97 N-terminus Domain and VBM Motif in Rhomboid Protease, RHBDL4 | | Descriptor: | Rhomboid-related protein 4, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Ly, T.T, Kang, J.Y, Lee, J.-G, An, J.Y, Youn, H.-S, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2015-11-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into the interaction of p97 N-terminus domain and VBM in rhomboid protease, RHBDL4.
Biochem.J., 473, 2016
|
|
5GLF
 
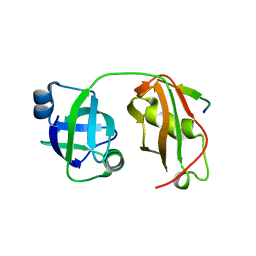 | | Structural insights into the interaction of p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease | | Descriptor: | Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Yoon, S.Y, Ly, T.T, Kang, J.Y, Youn, H.-S, An, J.Y, Lee, J.-G, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2016-07-11 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the interaction of human p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease.
FEBS Lett., 590, 2016
|
|
5I2L
 
 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
5I2Q
 
 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
