6LE5
 
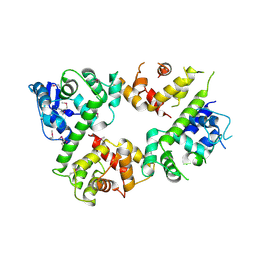 | | Crystal structure of the mitochondrial calcium uptake 1 and 2 heterodimer (MICU1-MICU2 heterodimer) in an apo state | | Descriptor: | Calcium uptake protein 1, mitochondrial, Calcium uptake protein 2 | | Authors: | Park, J, Lee, Y, Park, T, Kang, J.Y, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2019-11-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the MICU1-MICU2 heterodimer provides insights into the gatekeeping threshold shift.
Iucrj, 7, 2020
|
|
5E3X
 
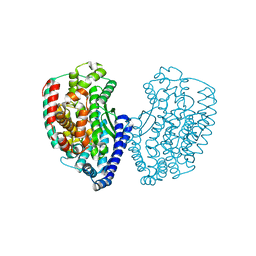 | | Crystal structure of thermostable Carboxypeptidase (FisCP) from Fervidobacterium Islandicum AW-1 | | Descriptor: | COBALT (II) ION, Thermostable carboxypeptidase 1 | | Authors: | Dhanasingh, I, Lee, Y.-J, Lee, D.W, Lee, S.H. | | Deposit date: | 2015-10-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Biochemical and structural characterization of a keratin-degrading M32 carboxypeptidase from Fervidobacterium islandicum AW-1
Biochem.Biophys.Res.Commun., 468, 2015
|
|
4R1Q
 
 | |
4R1O
 
 | |
1Q1Q
 
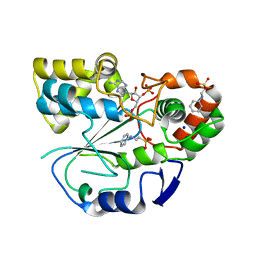 | | Crystal structure of human pregnenolone sulfotransferase (SULT2B1a) in the presence of PAP | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Lee, K.A, Fuda, H, Lee, Y.C, Negishi, M, Strott, C.A, Pedersen, L.C. | | Deposit date: | 2003-07-22 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone
and 3'-phosphoadenosine 5'-phosphate. Rationale for specificity differences between prototypical
SULT2A1 and the SULT2BG1 isoforms.
J.Biol.Chem., 278, 2003
|
|
1Q1Z
 
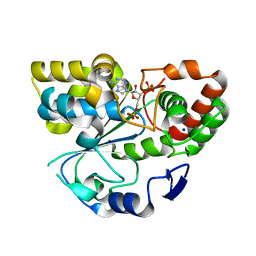 | | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, sulfotransferase family, ... | | Authors: | Lee, K.A, Fuda, H, Lee, Y.C, Negishi, M, Strott, C.A, Pedersen, L.C. | | Deposit date: | 2003-07-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone
and 3'-phosphoadenosine 5'-phosphate. Rationale for specificity differences between prototypical
SULT2A1 and the SULT2BG1 isoforms.
J.Biol.Chem., 278, 2003
|
|
1Q22
 
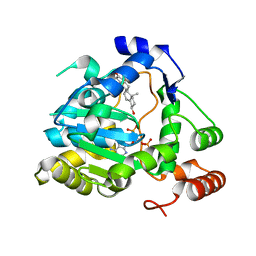 | | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of DHEA and PAP | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Lee, K.A, Fuda, H, Lee, Y.C, Negishi, M, Strott, C.A, Pedersen, L.C. | | Deposit date: | 2003-07-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone
and 3'-phosphoadenosine 5'-phosphate. Rationale for specificity differences between prototypical
SULT2A1 and the SULT2BG1 isoforms.
J.Biol.Chem., 278, 2003
|
|
1Q20
 
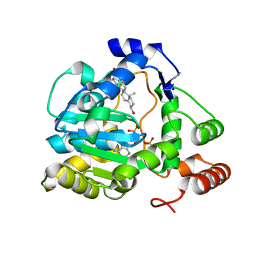 | | Crystal Structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of PAP and pregnenolone | | Descriptor: | (3BETA)-3-HYDROXYPREGN-5-EN-20-ONE, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Lee, K.A, Fuda, H, Lee, Y.C, Negishi, M, Strott, C.A, Pedersen, L.C. | | Deposit date: | 2003-07-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone
and 3'-phosphoadenosine 5'-phosphate. Rationale for specificity differences between prototypical
SULT2A1 and the SULT2BG1 isoforms.
J.Biol.Chem., 278, 2003
|
|
6GDG
 
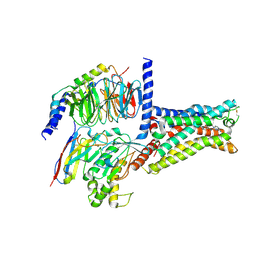 | | Cryo-EM structure of the adenosine A2A receptor bound to a miniGs heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Garcia-Nafria, J, Lee, Y. | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM structure of the adenosine A2Areceptor coupled to an engineered heterotrimeric G protein.
Elife, 7, 2018
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
7ENY
 
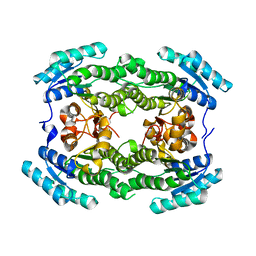 | | Crystal structure of hydroxysteroid dehydrogenase from Escherichia coli | | Descriptor: | 7alpha-hydroxysteroid dehydrogenase | | Authors: | Kim, K.-H, Lee, C.W, Pardhe, D.P, Hwang, J, Do, H, Lee, Y.M, Lee, J.H, Oh, T.-J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of an apo 7 alpha-hydroxysteroid dehydrogenase reveals key structural changes induced by substrate and co-factor binding.
J.Steroid Biochem.Mol.Biol., 212, 2021
|
|
8PM2
 
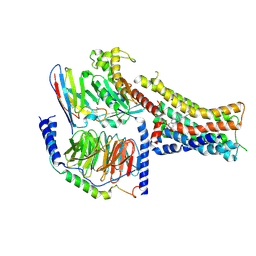 | | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gusach, A, Lee, Y, Edwards, P.C, Huang, F, Weyand, S.N, Tate, C.G. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f.
Biorxiv, 2023
|
|
8PVG
 
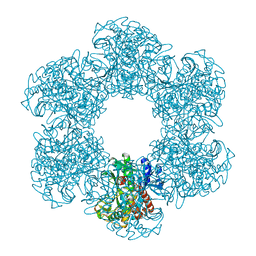 | | Structure of E. coli glutamine synthetase determined by cryoEM at 100 keV | | Descriptor: | Glutamine synthetase | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVC
 
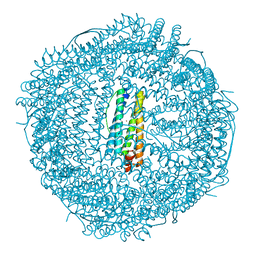 | | Structure of mouse heavy-chain apoferritin determined by cryoEM at 100 keV | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVE
 
 | | Structure of AHIR determined by cryoEM at 100 keV | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)) | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVJ
 
 | | Structure of lumazine synthase determined by cryoEM at 100 keV | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVD
 
 | | Structure of catalase determined by cryoEM at 100 keV | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PV9
 
 | | Structure of DPS determined by cryoEM at 100 keV | | Descriptor: | DNA protection during starvation protein | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVH
 
 | | Structure of human apo ALDH1A1 determined by cryoEM at 100 keV | | Descriptor: | Aldehyde dehydrogenase 1A1, CHLORIDE ION | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVB
 
 | | Structure of GABAAR determined by cryoEM at 100 keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVI
 
 | | Structure of PaaZ determined by cryoEM at 100 keV | | Descriptor: | Bifunctional protein PaaZ | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVF
 
 | | Structure of GAPDH determined by cryoEM at 100 keV | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G8W
 
 | | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | | Descriptor: | CARDIOLIPIN, GUANOSINE-5'-TRIPHOSPHATE, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Gogoi, P, Jones, S.A, Ruprecht, J.J, King, M.S, Lee, Y, Zogg, T, Pardon, E, Chand, D, Steimle, S, Copeman, D, Cotrim, C.A, Steyaert, J, Crichton, P.G, Moiseenkova-Bell, V, Kunji, E.R.S. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Sci Adv, 9, 2023
|
|
5I2Q
 
 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
