1R02
 
 | |
8T56
 
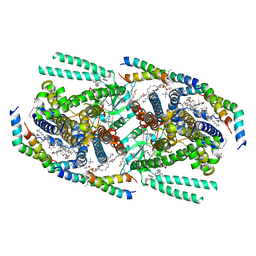 | | Structure of mechanically activated ion channel OSCA1.2 in peptidiscs | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Burendei, B, Jojoa-Cruz, S, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of mechanically activated ion channel OSCA2.3 reveals mobile elements in the transmembrane domain.
Structure, 32, 2024
|
|
7WJ5
 
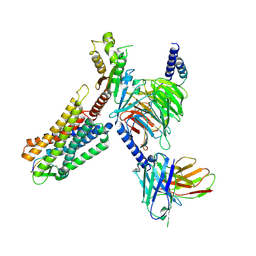 | | Cryo-EM structure of human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand binding specificity | | Descriptor: | Gai1 antibody (scfv16), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Heo, Y.S, Yoon, E.J, Jeon, Y.E, Yun, J.-H, Ishimoto, N, Woo, H, Park, S.Y, Song, J, Lee, W.T. | | Deposit date: | 2022-01-05 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of the human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand-binding specificity.
Elife, 11, 2022
|
|
8U53
 
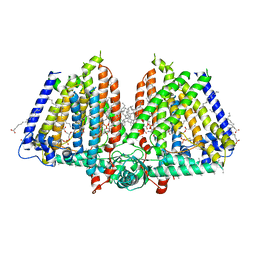 | | Mechanically activated ion channel OSCA3.1 in nanodiscs | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CSC1-like protein ERD4, PALMITIC ACID, ... | | Authors: | Jojoa-Cruz, S, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-guided mutagenesis of OSCAs reveals differential activation to mechanical stimuli.
Elife, 12, 2024
|
|
4LMQ
 
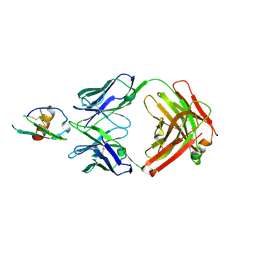 | | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 | | Descriptor: | Stromal cell-derived factor 1, hu30D8 Fab heavy chain, hu30D8 Fab light chain | | Authors: | Zhong, Z, Wang, J, Li, B, Xiang, H, Ultsch, M, Coons, M, Wong, T, Chiang, N.Y, Clark, S, Clark, R, Quintana, L, Gribling, P, Suto, E, Barck, K, Corpuz, R, Yao, J, Takkar, R, Lee, W.P, Damico-Beyer, L.A, Carano, R.D, Adams, C, Kelley, R.F, Wang, W, Ferrara, N. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12.
Clin.Cancer Res., 19, 2013
|
|
1V3Y
 
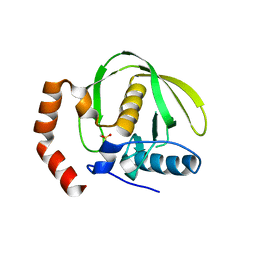 | | The crystal structure of peptide deformylase from Thermus thermophilus HB8 | | Descriptor: | Peptide deformylase | | Authors: | Kamo, M, Kudo, N, Lee, W.C, Ito, K, Motoshim, H, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-07 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The crystal structure of peptide deformylase from Thermus thermophilus HB8
to be published
|
|
1L3X
 
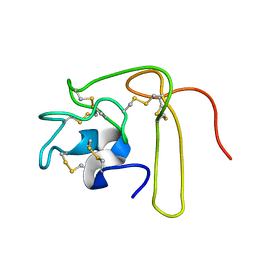 | | Solution Structure of Novel Disintegrin Salmosin | | Descriptor: | platelet aggregation inhibitor disintegrin | | Authors: | Shin, J, Lee, W. | | Deposit date: | 2002-03-01 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel disintegrin, salmosin, from Agkistrondon halys venom
Biochemistry, 42, 2003
|
|
2D7J
 
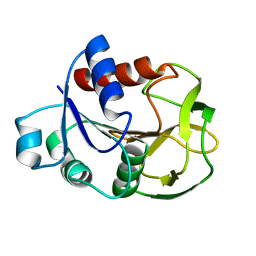 | | Crystal Structure Analysis of Glutamine Amidotransferase from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit A | | Authors: | Maruoka, S, Lee, W.C, Kamo, M, Kudo, N, Nagata, K, Tanokura, M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Pyrococcus horikoshii OT3
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
2D37
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D38
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D36
 
 | | The Crystal Structure of Flavin Reductase HpaC | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
6NF6
 
 | | Structure of chicken Otop3 in nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Otopetrin3 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NF4
 
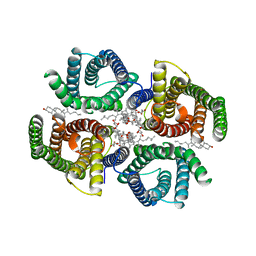 | | Structure of zebrafish Otop1 in nanodiscs | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, Otopetrin1 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2ABY
 
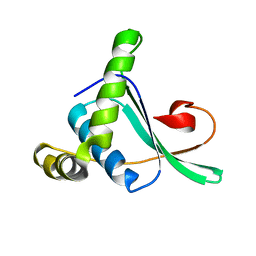 | | Solution structure of TA0743 from Thermoplasma acidophilum | | Descriptor: | hypothetical protein TA0743 | | Authors: | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
2EB1
 
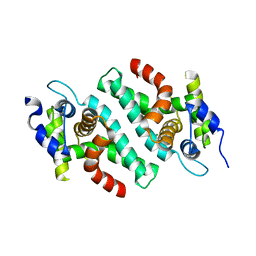 | | Crystal Structure of the C-Terminal RNase III Domain of Human Dicer | | Descriptor: | Endoribonuclease Dicer, MAGNESIUM ION | | Authors: | Takeshita, D, Zenno, S, Lee, W.C, Nagata, K, Saigo, K, Tanokura, M. | | Deposit date: | 2007-02-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homodimeric Structure and Double-stranded RNA Cleavage Activity of the C-terminal RNase III Domain of Human Dicer
J.Mol.Biol., 374, 2007
|
|
6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
5CHI
 
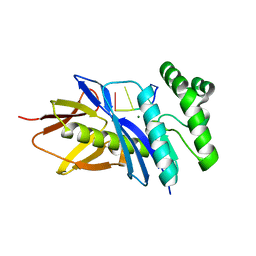 | | Crystal structure of PF2046 in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), MAGNESIUM ION, Uncharacterized protein | | Authors: | Kim, J.S, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural basis of two-nucleotide removal of ssDNA by a cryptic RNase H fold 3'-5' exonuclease PF2046 from Pyrococcus furiosus
to be published
|
|
8T57
 
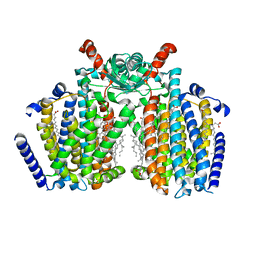 | | Structure of mechanically activated ion channel OSCA2.3 in peptidiscs | | Descriptor: | CHOLESTEROL, CSC1-like protein HYP1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Burendei, B, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of mechanically activated ion channel OSCA2.3 reveals mobile elements in the transmembrane domain.
Structure, 32, 2024
|
|
5CZY
 
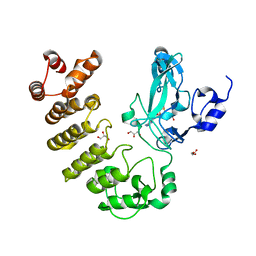 | | Crystal structure of LegAS4 | | Descriptor: | GLYCEROL, Legionella effector LegAS4, S-ADENOSYLMETHIONINE | | Authors: | Son, J, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Legionella pneumophila type IV secretion system effector LegAS4
Biochem.Biophys.Res.Commun., 465, 2015
|
|
2MPV
 
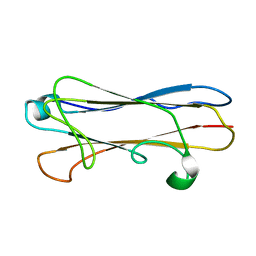 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
5TLC
 
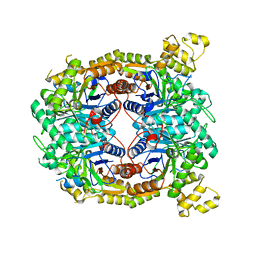 | |
4OH9
 
 | | Crystal Structure of the human MST2 SARAH homodimer | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2N7Y
 
 | | NMR structure of metal-binding domain 1 of ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Yu, C, Lee, W, Dmitriev, O. | | Deposit date: | 2015-09-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Metal Binding Domain 1 of the Copper Transporter ATP7B Reveals Mechanism of a Singular Wilson Disease Mutation.
Sci Rep, 8, 2018
|
|
4OH8
 
 | | Crystal Structure of the human MST1-RASSF5 SARAH heterodimer | | Descriptor: | Ras association domain-containing protein 5, Serine/threonine-protein kinase 4 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2G1E
 
 | | Solution Structure of TA0895 | | Descriptor: | hypothetical protein Ta0895 | | Authors: | Yeo, I.Y, Hong, E, Jung, J, Lee, W, Yee, A, Arrowsmith, C.H. | | Deposit date: | 2006-02-14 | | Release date: | 2006-12-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of TA0895, a MoaD homologue from Thermoplasma acidophilum
Proteins, 65, 2006
|
|
