5W18
 
 | | Staphylococcus aureus ClpP in complex with (S)-N-((2R,6S,8aS,14aS,20S,23aS)-2,6-dimethyl-5,8,14,19,23-pentaoxooctadecahydro-1H,5H,14H,19H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1]oxa[4,7,10,13]tetraazacyclohexadecin-20-yl)-3-phenyl-2-(3-phenylureido)propanamide | | Descriptor: | 9V7-PHE-SER-PRO-YCP-ALA-MP8, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ureadepsipeptides as ClpP Activators.
Acs Infect Dis., 2019
|
|
8E71
 
 | | Staphylococcus aureus ClpP in complex with compound 3471 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S,6S,9aS)-N-[(4-fluorophenyl)methyl]-6-methyl-8-[(naphthalen-1-yl)methyl]-4,7-dioxohexahydro-2H-pyrazino[1,2-a]pyrimidine-1(6H)-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Development of a high throughput and site specific, fluorescent polarization assay to screen for activators of Caseinolytic Protease P leads to the discovery of synthetically tractable new activator class
To Be Published
|
|
8E7P
 
 | | Staphylococcus aureus ClpP in complex with compound 3421 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5R,6S,9aS)-N-benzyl-6-[(4-hydroxyphenyl)methyl]-8-[(naphthalen-1-yl)methyl]-4,7-dioxohexahydro-2H-pyrazino[1,2-a]pyrimidine-1(6H)-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a high throughput and site specific, fluorescent polarization assay to screen for activators of Caseinolytic Protease P leads to the discovery of synthetically tractable new activator class
To Be Published
|
|
8EF8
 
 | | Staphylococcus aureus ClpP Y63W in complex with compound 3471 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S,6S,9aS)-N-[(4-fluorophenyl)methyl]-6-methyl-8-[(naphthalen-1-yl)methyl]-4,7-dioxohexahydro-2H-pyrazino[1,2-a]pyrimidine-1(6H)-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Development of a high throughput and site specific, fluorescent polarization assay to screen for activators of Caseinolytic Protease P leads to the discovery of synthetically tractable new activator class
To Be Published
|
|
6N80
 
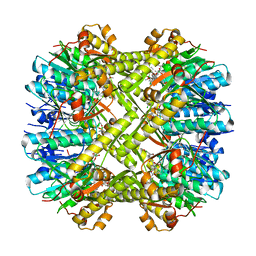 | | S. aureus ClpP bound to anti-4a | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, N-[(1R)-1-borono-3-methylbutyl]-N~2~-(2-chloro-4-methoxybenzene-1-carbonyl)-L-leucinamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-11-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | De Novo Design of Boron-Based Peptidomimetics as Potent Inhibitors of Human ClpP in the Presence of Human ClpX.
J.Med.Chem., 62, 2019
|
|
6CFD
 
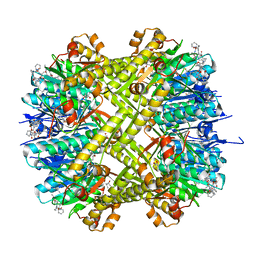 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
7JLS
 
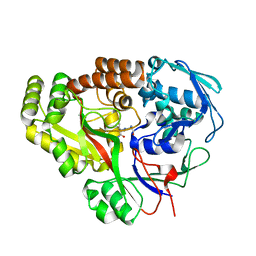 | | RV3666c bound to tripeptide | | Descriptor: | Peptide SER-VAL-ALA, Probable periplasmic dipeptide-binding lipoprotein DppA | | Authors: | Lee, R.E, Griffith, E.C, Miller, D.J. | | Deposit date: | 2020-07-30 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Biophysical analysis of the Mycobacteria tuberculosis peptide binding protein DppA reveals a stringent peptide binding pocket.
Tuberculosis (Edinb), 132, 2021
|
|
5N2M
 
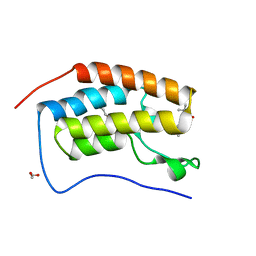 | | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, propan-2-yl ~{N}-[(2~{S},4~{R})-6-(3-acetamidophenyl)-1-ethanoyl-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
5N2L
 
 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, SULFATE ION, ... | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
6PKA
 
 | |
6PMD
 
 | |
9MVA
 
 | | Mycobacterium abscessus Eis2 in complex with Non-hydrolyzable CoenzymeA | | Descriptor: | 1,2-ETHANEDIOL, Amikacin resistance N-acetyltransferase Eis2, CHLORIDE ION, ... | | Authors: | Jayasinghe, T.D, Coker, A.L, Rajendra, T, Miller, D, Lee, R.E. | | Deposit date: | 2025-01-15 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium abscessus Eis2 in complex with Non-hydrolyzable CoenzymeA
To Be Published
|
|
9BC9
 
 | | Structure of KLHDC2 bound to SJ10278 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[2-chloro-4-(methoxymethoxy)phenyl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Principles of paralog-specific targeted protein degradation engaging the C-degron E3 KLHDC2.
Nat Commun, 15, 2024
|
|
9BCA
 
 | | Structure of KLHDC2 bound to SJ46411 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-{[2-(naphthalen-2-yl)-1,3-thiazol-4-yl]acetyl}glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of paralog-specific targeted protein degradation engaging the C-degron E3 KLHDC2.
Nat Commun, 15, 2024
|
|
9BCC
 
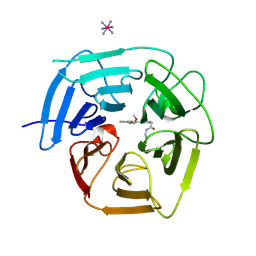 | | Structure of KLHDC2 bound to SJ46418 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[8-(2-methoxyethoxy)naphthalen-2-yl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of paralog-specific targeted protein degradation engaging the C-degron E3 KLHDC2.
Nat Commun, 15, 2024
|
|
5VZ2
 
 | |
8UVS
 
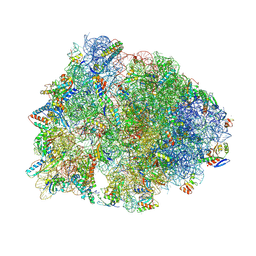 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin derivative 2694, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.75A resolution | | Descriptor: | (2R,4R,4aS,5aR,6S,7S,8R,9S,9aR,10aS)-2-methyl-6,8-bis(methylamino)-4-({[2-(oxan-4-yl)ethyl]amino}methyl)octahydro-2H-pyrano[2,3-b][1,4]benzodioxine-4,4a,7,9(10aH)-tetrol, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UVR
 
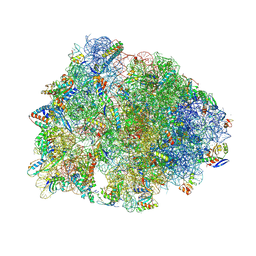 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7UY4
 
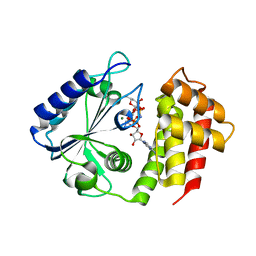 | | Aminoglycoside-modifying enzyme ANT-3,9 in complex with spectinomycin and AMP-PNP | | Descriptor: | Aminoglycoside (3'') (9) adenylyltransferase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Reeve, S.M, Lee, R.E, Das, N, Jayaraman, S. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Characterization of the spectinomycin deactivating enzyme ANT-3,9
To Be Published
|
|
7UES
 
 | | PANK3 complex structure with compound PZ-4202 | | Descriptor: | MAGNESIUM ION, N-(4-{2-[4-(6-cyanopyridazin-3-yl)piperazin-1-yl]-2-oxoethyl}phenyl)methanesulfonamide, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | White, S.W, Yun, M, Lee, R.E. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of hPANK Activators with Improved Pharmacological Properties
To Be Published
|
|
7UER
 
 | |
7UEV
 
 | |
7UEQ
 
 | | PANK3 complex structure with compound PZ-4061 | | Descriptor: | 1-[4-(6-chloropyridazin-3-yl)piperazin-1-yl]-2-[4-(dimethylamino)phenyl]ethan-1-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | White, S.W, Yun, M, Lee, R.E. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Brain Penetrant Pyridazine Pantothenate Kinase Activators.
J.Med.Chem., 67, 2024
|
|
7UET
 
 | | PANK3 complex structure with compound PZ-4140 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-(4-{2-[4-(6-chloropyridazin-3-yl)piperazin-1-yl]-2-oxoethyl}phenyl)methanesulfonamide, ... | | Authors: | White, S.W, Yun, M, Lee, R.E. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of hPANK Activators with Improved Pharmacological Properties
To Be Published
|
|
7UEY
 
 | | PANK3 complex structure with compound PZ-4128 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-(4-{2-[4-(6-chloropyridazin-3-yl)piperazin-1-yl]-2-oxoethyl}phenyl)-2-hydroxyacetamide, ... | | Authors: | White, S.W, Yun, M, Lee, R.E. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of hPANK Activators with Improved Pharmacological Properties
To Be Published
|
|
