4QIU
 
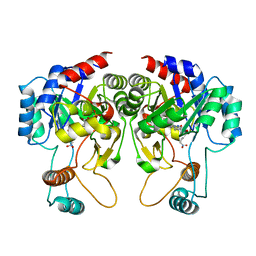 | |
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
8GJE
 
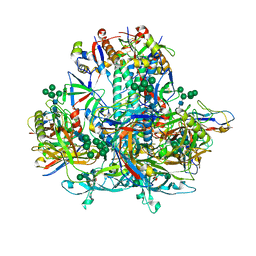 | | HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC117 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, J.H, Ward, A.B. | | Deposit date: | 2023-03-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan heterogeneity as a cause of the persistent fraction in HIV-1 neutralization.
Plos Pathog., 19, 2023
|
|
5UY3
 
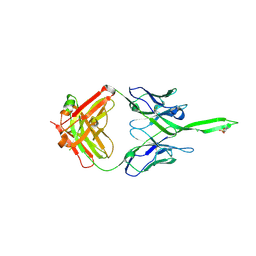 | | Crystal structure of human Fab PGT144, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Antibody PGT144 Fab heavy chain, Antibody PGT144 Fab light chain | | Authors: | Julien, J.-P, Lee, J.H, Wilson, I.A. | | Deposit date: | 2017-02-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
6LVP
 
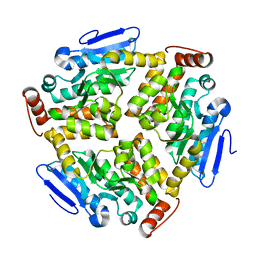 | |
6LVO
 
 | |
4ZZ7
 
 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
5AYX
 
 | | Crystal structure of Human Quinolinate Phosphoribosyltransferase | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Kang, G.B, Kim, M.-K, Im, Y.J, Lee, J.H, Youn, H.-S, An, J.Y, Lee, J.-G, Fukuoka, S.-I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
6L3A
 
 | | Cytochrome P450 107G1 (RapN) with everolimus | | Descriptor: | Cytochrome P450, Everolimus, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Km, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
6L39
 
 | | Cytochrome P450 107G1 (RapN) | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Kim, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
7VPF
 
 | |
7XJT
 
 | |
5H1Z
 
 | | CYP153D17 from Sphingomonas sp. PAMC 26605 | | Descriptor: | DODECANE, PROTOPORPHYRIN IX CONTAINING FE, putative CYP alkane hydroxylase CYP153D17 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Putative Cytochrome P450 Alkane Hydroxylase (CYP153D17) from Sphingomonas sp. PAMC 26605 and Its Conformational Substrate Binding
Int J Mol Sci, 17, 2016
|
|
5IT1
 
 | | Streptomyces peucetius CYP105P2 complex with biphenyl compound | | Descriptor: | 4,4'-PROPANE-2,2-DIYLDIPHENOL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-03-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450 (CYP105P2) from Streptomyces peucetius and Its Conformational Changes in Response to Substrate Binding
Int J Mol Sci, 17, 2016
|
|
3EGM
 
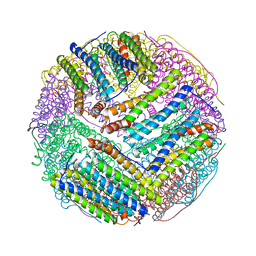 | | Structural basis of iron transport gating in Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Shin, H.J, Lee, J.H. | | Deposit date: | 2008-09-11 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of ferritin from Helicobacter pylori reveals unusual conformational changes for iron uptake.
J.Mol.Biol., 390, 2009
|
|
7XRI
 
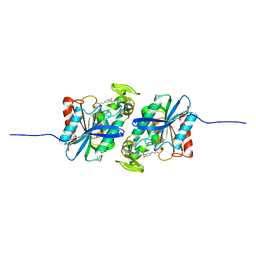 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7YVT
 
 | |
7XRH
 
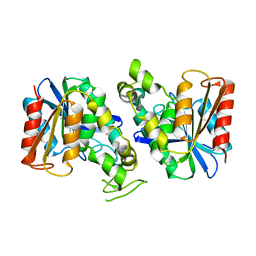 | | Feruloyl esterase from Lactobacillus acidophilus | | Descriptor: | Cinnamoyl esterase | | Authors: | Hwang, J, Lee, C.W, Lee, J.H, Do, H. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
3BWK
 
 | | Crystal Structure of Falcipain-3 with Its inhibitor, K11017 | | Descriptor: | Cysteine protease falcipain-3, N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide, SULFATE ION | | Authors: | Kerr, I, Lee, J.H, Brinen, L.S. | | Deposit date: | 2008-01-09 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Vinyl sulfones as antiparasitic agents and a structural basis for drug design.
J.Biol.Chem., 284, 2009
|
|
5Y7W
 
 | | Crystal structure of the Nco-A1 PAS-B domain with YL-2 | | Descriptor: | Nuclear receptor coactivator 1, YL-2 peptide | | Authors: | Lee, Y.J, Yoon, H.S, Lee, J.H, Bae, J.H, Song, J.Y, Lim, H.S. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeted Inhibition of the NCOA1/STAT6 Protein-Protein Interaction
J. Am. Chem. Soc., 139, 2017
|
|
2ZIW
 
 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIU
 
 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
7X99
 
 | |
2ZIV
 
 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
