2KZN
 
 | | Solution NMR Structure of Peptide methionine sulfoxide reductase msrB from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR10 | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Ertekin, A, Maglaqui, M, Janjua, H, Cooper, B, Ciccosanti, C, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J, Lee, H, Aramini, J.M, Rossi, P, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UVM
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL4 | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVO
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1B | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVN
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVK
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase MLL2, SULFATE ION, ... | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVL
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL3 | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
2LF0
 
 | | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering, NESG target SfR339/OCSP target sf3636 | | Descriptor: | Uncharacterized protein yibL | | Authors: | Wu, B, Lemak, A, Yee, A, Lee, H, Gutmanas, A, Semesi, A, Garcia, M, Fang, X, Wang, Y, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering
To be Published
|
|
2LE1
 
 | | Solution NMR Structure of Tfu_2981 from Thermobifida fusca, Northeast Structural Genomics Consortium Target TfR85A | | Descriptor: | Uncharacterized protein | | Authors: | Pulavarti, S.V.S.R.K, Eletsky, A, Mills, J.L, Sukumaran, D.K, Wang, D, Ciccosanti, C, Hamilton, K, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-03 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Tfu_2981 from Thermobifida fusca, Northeast Structural Genomics Consortium Target TfR85A
To be Published
|
|
2JQL
 
 | | NMR structure of the yeast Dun1 FHA domain in complex with a doubly phosphorylated (pT) peptide derived from Rad53 SCD1 | | Descriptor: | DNA damage response protein kinase DUN1, Serine/threonine-protein kinase RAD53 | | Authors: | Yuan, C, Lee, H, Chang, C, Heierhorst, J, Tsai, M. | | Deposit date: | 2007-06-02 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade.
Mol.Cell, 30, 2008
|
|
2JQJ
 
 | | NMR structure of yeast Dun1 FHA domain | | Descriptor: | DNA damage response protein kinase DUN1 | | Authors: | Yuan, C, Lee, H, Chang, C, Heierhorst, J, Tsai, M. | | Deposit date: | 2007-06-02 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade.
Mol.Cell, 30, 2008
|
|
2KD1
 
 | | Solution NMR structure of the integrase-like domain from Bacillus cereus ordered locus BC_1272. Northeast Structural Genomics Consortium Target BcR268F | | Descriptor: | DNA integration/recombination/invertion protein | | Authors: | Rossi, P, Lee, H, Maglaqui, M, Foote, E.L, Buchwald, W.A, Jiang, M, Swapna, G.V.T, Nair, R, Xiao, R, Acton, T.B, Rost, B, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-31 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Integrase-Like Domain from Bacillus cereus Ordered Locus BC_1272. Northeast Structural Genomics Consortium Target BcR268F.
To be Published
|
|
2KJ5
 
 | | Solution NMR structure of a domain from a putative phage integrase protein Nmul_A0064 from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR46C | | Descriptor: | Phage integrase | | Authors: | Mills, J.L, Eletsky, A, Ghosh, A, Wang, D, Lee, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a domain from a putative phage integrase protein Nmul_A0064 from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR46C.
To be Published
|
|
2KSO
 
 | | EphA2:SHIP2 SAM:SAM complex | | Descriptor: | Ephrin type-A receptor 2, Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Hota, P.K, Preeti, C, Stetzik, L, Kim, S, Wang, B, Lee, H, Buck, M. | | Deposit date: | 2010-01-10 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the EphA2:SHIP2 SAM domain heterodimer
To be Published
|
|
2HNU
 
 | | Crystal Structure of a Dipeptide Complex of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2HNV
 
 | | Crystal Structure of a Dipeptide Complex of the Q58V Mutant of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
5T1V
 
 | | Crystal structure of Zika virus NS2B-NS3 protease in apo-form. | | Descriptor: | NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in apo-form.
To Be Published
|
|
2HNW
 
 | | Crystal Structure of the F91STOP mutant of des1-6 Bovine Neurophysin-I, unliganded state | | Descriptor: | Oxytocin-neurophysin 1 | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2HEM
 
 | | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit. | | Descriptor: | 5'-R(P*GP*GP*GP*AP*AP*GP*GP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*CP*GP*GP*CP*CP*C)-3' | | Authors: | Zhao, Q, Nagaswamy, U, Lee, H, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2006-06-21 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit
Nucleic Acids Res., 33, 2005
|
|
2KZ6
 
 | | Solution structure of protein CV0426 from Chromobacterium violaceum, Northeast structural genomics consortium (NESG) target CVT2 | | Descriptor: | Uncharacterized protein | | Authors: | Lemak, A, Yee, A, Lee, H, Semesi, A, Prestegard, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-11 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein CV0426 from Chromobacterium violaceum
To be Published
|
|
2KYY
 
 | | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea, Northeast Structural Genomics Consortium Target NeR70A | | Descriptor: | Possible ATP-dependent DNA helicase RecG-related protein | | Authors: | Eletsky, A, Mills, J.L, Lee, H, Maglaqui, M, Ciccosanti, C, Hamilton, K, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea
To be Published
|
|
2KXY
 
 | | Solution structure of SuR18C from Streptococcus thermophilus. Northeast Structural Genomics Consortium Target SuR18C | | Descriptor: | Uncharacterized protein | | Authors: | Liu, Y, Lee, H, Belote, R, Ciccosanti, C, Hamilton, K, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SuR18C from Streptococcus thermophilus. Northeast Structural Genomics Consortium Target SuR18C
To be Published
|
|
2L02
 
 | | Solution NMR Structure of protein BT2368 from Bacteroides thetaiotaomicron, Northeast Structural Genomics Consortium Target BtR375 | | Descriptor: | Uncharacterized protein | | Authors: | Eletsky, A, Lee, H, Wang, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of protein BT2368 from Bacteroides thetaiotaomicron
To be Published
|
|
3FCA
 
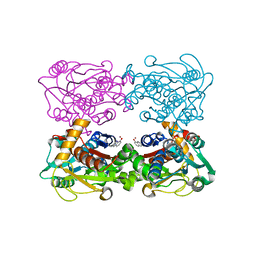 | | Genetic Incorporation of a Metal-ion Chelating Amino Acid into proteins as biophysical probe | | Descriptor: | Cysteine synthase, ZINC ION | | Authors: | Wang, F, Lee, H, Spraggon, G, Schultz, P.G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Genetic incorporation of a metal-ion chelating amino acid into proteins as a biophysical probe.
J.Am.Chem.Soc., 131, 2009
|
|
6JRP
 
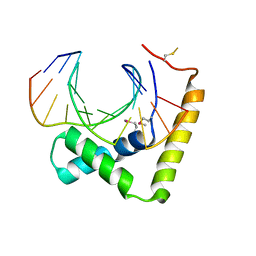 | | Crystal structure of CIC-HMG-ETV5-DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*AP*AP*TP*GP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*CP*AP*TP*TP*CP*AP*T)-3'), Protein capicua homolog | | Authors: | Song, J.J, Lee, H. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-31 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of Capicua HMG-box domain complexed with the ETV5-DNA and its implications for Capicua-mediated cancers.
Febs J., 286, 2019
|
|
6KWA
 
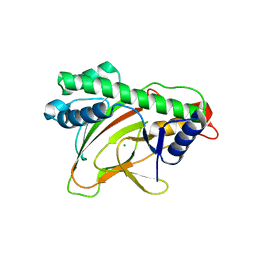 | |
