7MFT
 
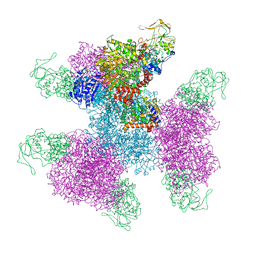 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex (GudB6-GltA6-GltB6) | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-11 | | Release date: | 2022-01-05 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
8E41
 
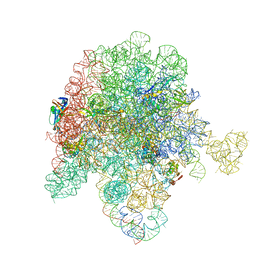 | | E. coli 50S ribosome bound to tiamulin and VS1 | | Descriptor: | 50S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Tiamulin and NPET binders
To Be Published
|
|
8E3L
 
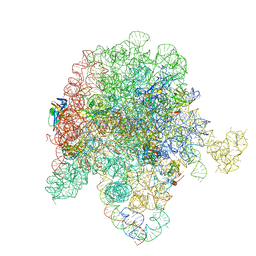 | | E. coli 50S ribosome bound to D-linker solithromycin conjugate | | Descriptor: | (2~{R})-~{N}-[(2~{R})-6-azanyl-1-[[(2~{R})-1-[[(2~{R})-1-[[3-[1-[4-[(1~{S},2~{R},5~{S},7~{R},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-5-fluoranyl-9-methoxy-1,5,7,11,13-pentamethyl-4,6,12,16-tetrakis(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-15-yl]butyl]-1,2,3-triazol-4-yl]phenyl]amino]-4-methylsulfanyl-1-oxidanylidene-butan-2-yl]amino]-3-(4-hydroxyphenyl)-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-hexan-2-yl]-1-[(2~{R})-2-[[(2~{R})-2-[2-[[2,3-bis(oxidanyl)phenyl]carbonyl-[4-[[2,3-bis(oxidanyl)phenyl]carbonylamino]butyl]amino]ethanoylamino]-3-(1~{H}-indol-3-yl)propanoyl]amino]-3-oxidanyl-propanoyl]pyrrolidine-2-carboxamide, 50S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Solithromycin siderophore conjugates
To Be Published
|
|
8E47
 
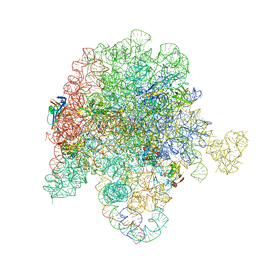 | | E. coli 50S ribosome bound to antibiotic analog SLC26 | | Descriptor: | (2R,3S,4R,5R,8R,10R,11R,12S,13S,14R)-2-ethyl-3,4,10-trihydroxy-3,5,6,8,10,12,14-heptamethyl-15-oxo-11-({3,4,6-trideoxy-3-[(2-{[2-({[(5S)-3-{(4M)-3-fluoro-4-[(6P)-6-(2-methyl-2H-tetrazol-5-yl)pyridin-3-yl]phenyl}-2-oxo-1,3-oxazolidin-5-yl]methyl}amino)-2-oxoethyl]sulfanyl}ethyl)(methyl)amino]-beta-D-xylo-hexopyranosyl}oxy)-1-oxa-6-azacyclopentadecan-13-yl 2,6-dideoxy-3-C-methyl-3-O-methyl-alpha-L-ribo-hexopyranoside, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | SLC collection of antibiotic analogs
To Be Published
|
|
8E46
 
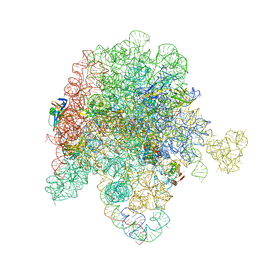 | | E. coli 50S ribosome bound to antibiotic analog SLC21 | | Descriptor: | (2R,3S,4R,5R,8R,10R,11R,12S,13S,14R)-2-ethyl-3,4,10-trihydroxy-3,5,6,8,10,12,14-heptamethyl-15-oxo-11-({3,4,6-trideoxy-3-[{[1-(2-{[(1R,2R)-1,3-dihydroxy-1-(4-nitrophenyl)propan-2-yl]amino}-2-oxoethyl)-1H-1,2,3-triazol-4-yl]methyl}(methyl)amino]-beta-D-xylo-hexopyranosyl}oxy)-1-oxa-6-azacyclopentadecan-13-yl 2,6-dideoxy-3-C-methyl-3-O-methyl-alpha-L-ribo-hexopyranoside, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | SLC collection of antibiotic analogs
To Be Published
|
|
2PX7
 
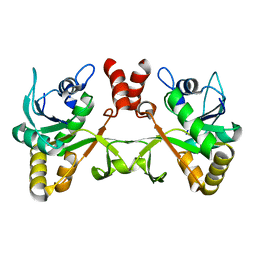 | | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | | Authors: | Chen, L, Tsukuda, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Chen, L.-Q, Liu, Z.-J, Lee, D, Chang, S.-H, Nguyen, D, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-14 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8.
To be Published
|
|
4B04
 
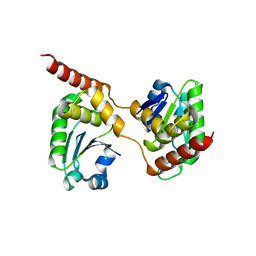 | | Crystal structure of the Catalytic Domain of Human DUSP26 (C152S) | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 26 | | Authors: | Won, E.-Y, Lee, D.Y, Park, S.G, Yokoyama, S, Kim, S.J, Chi, S.-W. | | Deposit date: | 2012-06-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | High-Resolution Crystal Structure of the Catalytic Domain of Human Dual-Specificity Phosphatase 26
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1TEX
 
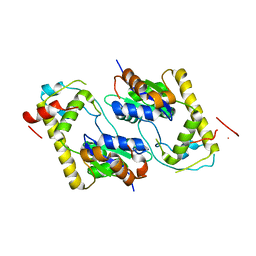 | | Mycobacterium smegmatis Stf0 Sulfotransferase with Trehalose | | Descriptor: | Stf0 Sulfotransferase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Mougous, J.D, Petzold, C.J, Senaratne, R.H, Lee, D.H, Akey, D.L, Lin, F.L, Munchel, S.E, Pratt, M.R, Riley, L.W, Leary, J.A, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2004-05-25 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis.
Nat.Struct.Mol.Biol., 11, 2004
|
|
8E33
 
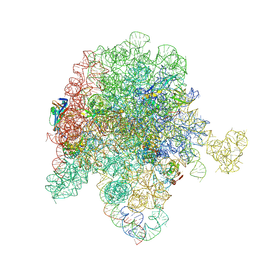 | | E. coli 50S ribosome bound to compound streptogramin analog SAB001 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E43
 
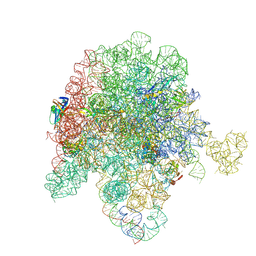 | | E. coli 50S ribosome bound to compound streptogramin A analog 3336 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,23S,26aR)-16-fluoro-14-hydroxy-12-methyl-1,7,22-trioxo-4-(prop-2-en-1-yl)-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Seiple, I.B, Fraser, J.S. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Streptogramin A analogs
To Be Published
|
|
8E44
 
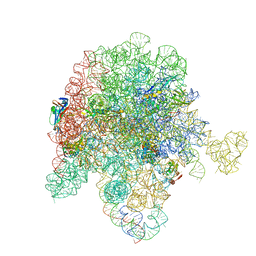 | | E. coli 50S ribosome bound to antibiotic analog SLC09 | | Descriptor: | 2-chloro-N-{[(5S)-3-{(4M)-3-fluoro-4-[(6P)-6-(2-methyl-2H-tetrazol-5-yl)pyridin-3-yl]phenyl}-2-oxo-1,3-oxazolidin-5-yl]methyl}acetamide, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | SLC collection of antibiotic analogs
To Be Published
|
|
8E45
 
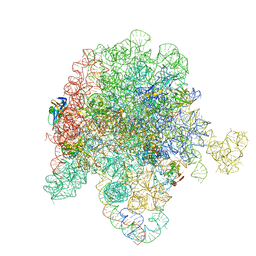 | | E. coli 50S ribosome bound to antibiotic analog SLC17 | | Descriptor: | 2-{[(2S,3R,4S,6R)-2-{[(2R,3S,4R,5R,8R,10R,11R,12S,13S,14R)-2-ethyl-3,4,10-trihydroxy-13-{[(2R,4R,5S,6S)-5-hydroxy-4-methoxy-4,6-dimethyloxan-2-yl]oxy}-3,5,6,8,10,12,14-heptamethyl-15-oxo-1-oxa-6-azacyclopentadecan-11-yl]oxy}-3-hydroxy-6-methyloxan-4-yl](methyl)amino}-N-{[(5S)-3-{(4M)-3-fluoro-4-[(6P)-6-(2-methyl-2H-tetrazol-5-yl)pyridin-3-yl]phenyl}-2-oxo-1,3-oxazolidin-5-yl]methyl}acetamide (non-preferred name), 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | SLC collection of antibiotic analogs
To Be Published
|
|
8E30
 
 | | E. coli 50S ribosome bound to compound streptogramin A analog 3142 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (1.91 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E32
 
 | | E. coli 50S ribosome bound to compound streptogramin analogs SA1 and SB1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,23S,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-yn-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E48
 
 | | E. coli 50S ribosome bound to antibiotic analog SLC30 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | SLC collection of antibiotic analogs
To Be Published
|
|
8E36
 
 | | E. coli 50S ribosome bound to compound streptogramin A analog 3146 | | Descriptor: | 2-[(3R,4R,5E,10E,12E,14S,16R,23S,26aR)-16-fluoro-14-hydroxy-12-methyl-1,7,22-trioxo-3-(propan-2-yl)-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-4-yl]-N-{2-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]ethyl}acetamide, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E42
 
 | | E. coli 50S ribosome bound to tiamulin and azithromycin | | Descriptor: | 50S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Tiamulin and NPET binders
To Be Published
|
|
8E49
 
 | | E. coli 50S ribosome bound to antibiotic analog SLC31 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | SLC collection of antibiotic analogs
To Be Published
|
|
8E35
 
 | | E. coli 50S ribosome bound to compound SAB002 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
6KGZ
 
 | | bacterial cystathionine gamma-lyase MccB of Staphylococcus aureus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cystathionine gamma-lyase | | Authors: | Ha, N.-C, Lee, D. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Bacterial Cystathionine Gamma-Lyase in The Cysteine Biosynthesis Pathway of Staphylococcus aureus
Crystals, 9, 2019
|
|
2WXI
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with SW30. | | Descriptor: | 2-{[4-amino-3-(3-hydroxyprop-1-yn-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-5-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, GLYCEROL, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXK
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with INK666. | | Descriptor: | 3-(2-amino-1,3-benzothiazol-6-yl)-1-{[2-(4-methylpiperazin-1-yl)quinolin-3-yl]methyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXN
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with DL07. | | Descriptor: | 3-{[4-amino-1-(1-methylethyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]ethynyl}phenol, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXH
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with SW14. | | Descriptor: | 2-{[4-amino-3-(3-fluoro-4-hydroxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-5-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXG
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with SW13. | | Descriptor: | 2-{[4-amino-3-(3-fluoro-5-hydroxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-5-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
