1AQC
 
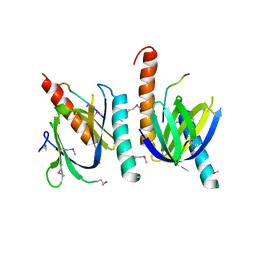 | | X11 PTB DOMAIN-10MER PEPTIDE COMPLEX | | Descriptor: | PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1997-12-24 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
8IDR
 
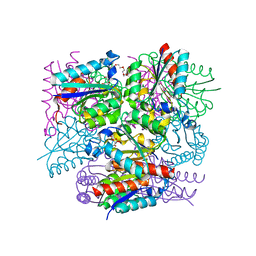 | |
8WZX
 
 | | Cryo-EM structure of the hamster prion 23-144 fibril at pH 3.7 | | Descriptor: | Major prion protein | | Authors: | Lee, C.-H, Saw, J.-E, Chen, E, Wang, C.-H, Chen, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The Positively Charged Cluster in the N-terminal Disordered Region may Affect Prion Protein Misfolding: Cryo-EM Structure of Hamster PrP(23-144) Fibrils.
J.Mol.Biol., 436, 2024
|
|
6A4A
 
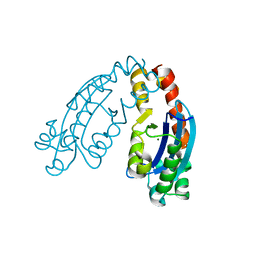 | |
6A4D
 
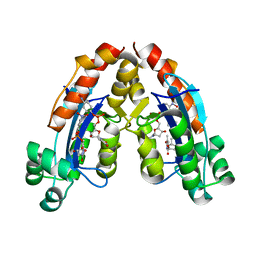 | |
6A7I
 
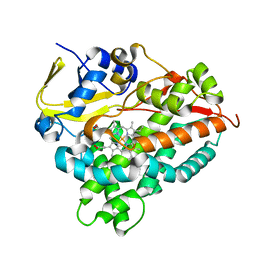 | | CYP154C4 from Streptomyces sp. W2061 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
6A4F
 
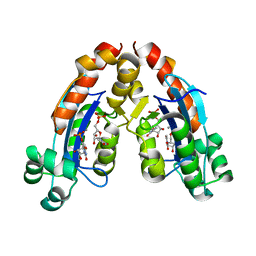 | |
6A7J
 
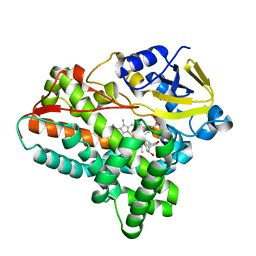 | | Testerone bound CYP154C4 from Streptomyces sp. ATCC 11861 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, TESTOSTERONE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
4BWZ
 
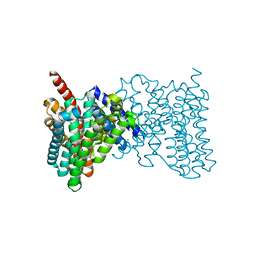 | |
4Z55
 
 | |
6JZL
 
 | |
8IDU
 
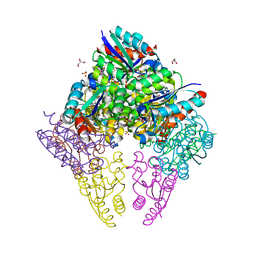 | | Crystal structure of substrate bound-form dehydroquinate dehydratase from Corynebacterium glutamicum | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL, ... | | Authors: | Lee, C.H, Kim, S, Kim, K.-J. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Analysis of 3-Dehydroquinate Dehydratase from Corynebacterium glutamicum .
J Microbiol Biotechnol., 33, 2023
|
|
6A4E
 
 | |
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
5U6O
 
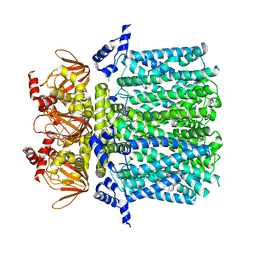 | |
5DRO
 
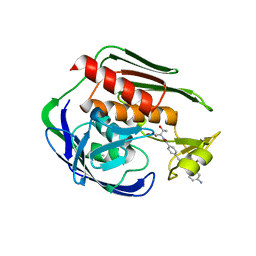 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRR
 
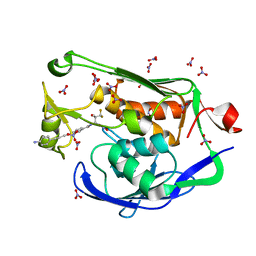 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-058 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRQ
 
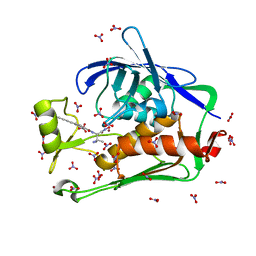 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-040 complex | | Descriptor: | N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5BUG
 
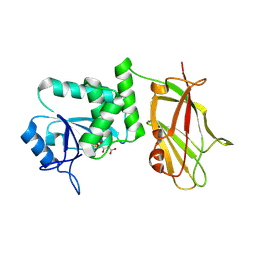 | | Crystal structure of human phosphatase PTEN oxidized by H2O2 | | Descriptor: | L(+)-TARTARIC ACID, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Lee, C.-U, Bier, D, Hennig, S, Grossmann, T.N. | | Deposit date: | 2015-06-03 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Redox Modulation of PTEN Phosphatase Activity by Hydrogen Peroxide and Bisperoxidovanadium Complexes.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1X11
 
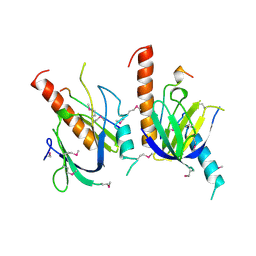 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
3L9P
 
 | |
4LCG
 
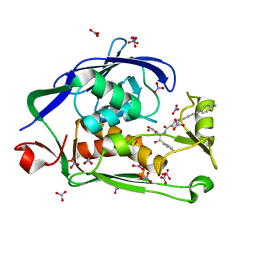 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-050 complex | | Descriptor: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.568 Å) | | Cite: | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4IS9
 
 | | Crystal Structure of the Escherichia coli LpxC/L-161,240 complex | | Descriptor: | (4R)-2-(3,4-dimethoxy-5-propylphenyl)-N-hydroxy-4,5-dihydro-1,3-oxazole-4-carboxamide, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
4MQY
 
 | | Crystal Structure of the Escherichia coli LpxC/LPC-138 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-2-methyl-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
5U6P
 
 | |
