7PPJ
 
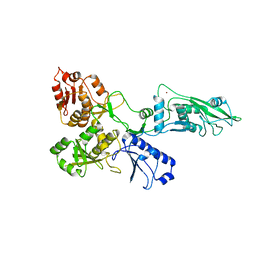 | | human SLFN5 | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Lammens, K, Metzner, F.J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 50, 2022
|
|
8S86
 
 | | human PLD3 homodimer structure | | Descriptor: | 5'-3' exonuclease PLD3 | | Authors: | Lammens, K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation.
Immunity, 57, 2024
|
|
3QG5
 
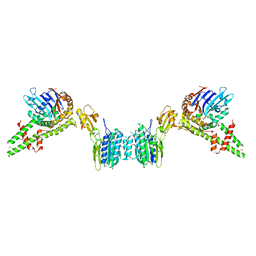 | |
7ZQY
 
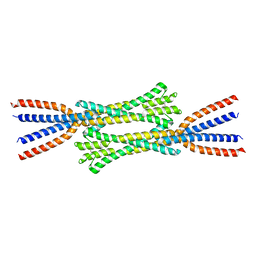 | | Chaetomium thermophilum Rad50 Zn hook | | Descriptor: | DH domain-containing protein, ZINC ION | | Authors: | Lammens, K, Rotheneder, M, Stakyte, K. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
4W9M
 
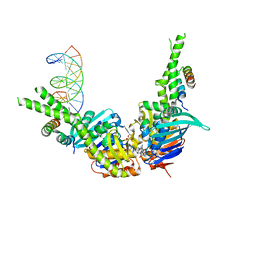 | | AMPPNP bound Rad50 in complex with dsDNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*GP*TP*CP*AP*CP*CP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*GP*GP*TP*GP*AP*CP*CP*GP*AP*CP*C)-3'), Exonuclease, ... | | Authors: | Rojowska, A, Lammens, K. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Rad50 DNA double-strand break repair protein in complex with DNA.
Embo J., 33, 2014
|
|
4YKE
 
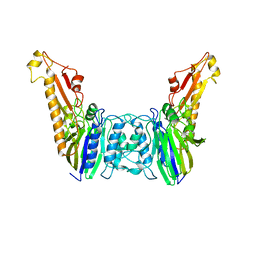 | |
2QFD
 
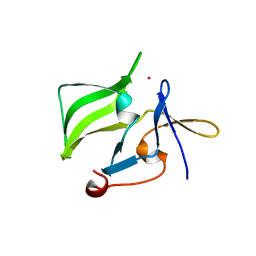 | | Crystal structure of the regulatory domain of human RIG-I with bound Hg | | Descriptor: | MERCURY (II) ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Cui, S, Lammens, A, Lammens, K, Hopfner, K.P. | | Deposit date: | 2007-06-27 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I.
Mol.Cell, 29, 2008
|
|
2QFB
 
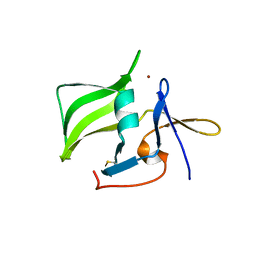 | | Crystal structure of the regulatory domain of human RIG-I with bound Zn | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, ZINC ION | | Authors: | Cui, S, Lammens, A, Lammens, K, Hopfner, K.P. | | Deposit date: | 2007-06-27 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I.
Mol.Cell, 29, 2008
|
|
2W4R
 
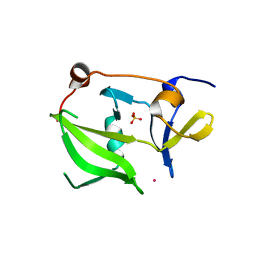 | | Crystal structure of the regulatory domain of human LGP2 | | Descriptor: | MERCURY (II) ION, PROBABLE ATP-DEPENDENT RNA HELICASE DHX58, SULFATE ION | | Authors: | Pippig, D.A, Hellmuth, J.C, Cui, S, Kirchhofer, A, Lammens, K, Lammens, A, Schmidt, A, Rothenfusser, S, Hopfner, K.P. | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Regulatory Domain of the Rig-I Family ATPase Lgp2 Senses Double-Stranded RNA.
Nucleic Acids Res., 37, 2009
|
|
7A08
 
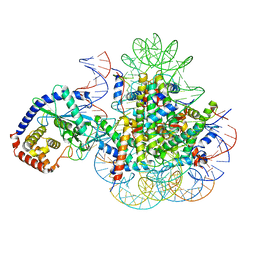 | | CryoEM Structure of cGAS Nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Michalski, S, de Oliveira Mann, C.C, Witte, G, Bartho, J, Lammens, K, Hopfner, K.P. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for sequestration and autoinhibition of cGAS by chromatin.
Nature, 587, 2020
|
|
9ERE
 
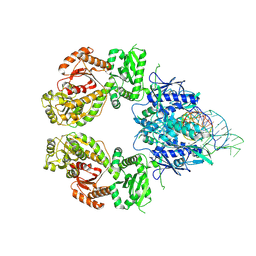 | | SLFN11 dimer bound to tRNA-Leu-TAA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, RNA (5'-R(P*CP*CP*UP*GP*GP*UP*AP*CP*CP*A)-3'), ... | | Authors: | Kugler, M, Metzner, F.J, Lammens, K. | | Deposit date: | 2024-03-22 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Phosphorylation-mediated conformational change regulates human SLFN11.
Nat Commun, 15, 2024
|
|
9ERF
 
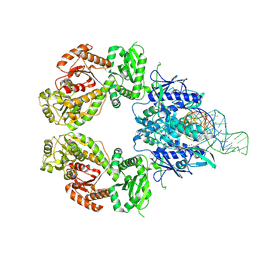 | | SLFN11 dimer bound to tRNA-Met-CAT | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, RNA (5'-R(P*UP*CP*UP*GP*CP*UP*AP*CP*CP*A)-3'), ... | | Authors: | Kugler, M, Metzner, F.J, Lammens, K. | | Deposit date: | 2024-03-22 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Phosphorylation-mediated conformational change regulates human SLFN11.
Nat Commun, 15, 2024
|
|
9ERD
 
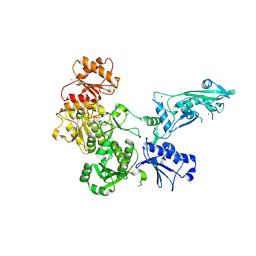 | | Human SLFN11 S753D monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Schlafen family member 11, ... | | Authors: | Kugler, M, Metzner, F.J, Lammens, K. | | Deposit date: | 2024-03-22 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Phosphorylation-mediated conformational change regulates human SLFN11.
Nat Commun, 15, 2024
|
|
2XGP
 
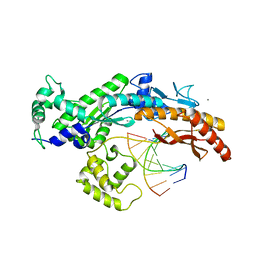 | | Yeast DNA polymerase eta in complex with C8-2-acetylaminofluorene containing DNA | | Descriptor: | 5'-D(*CP*8FG*CP*TP*CP*AP*TP*CP*CP*AP*C)-3', 5'-D(*GP*TP*GP*GP*AP*TP*GP*AP*G)-3', CALCIUM ION, ... | | Authors: | Scheider, S, Lammens, K, Schorr, S, Hopfner, K.P, Carell, T. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Replication Blocking and Bypass of Y-Family Polymerase Eta by Bulky Acetylaminofluorene DNA Adducts.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8BAH
 
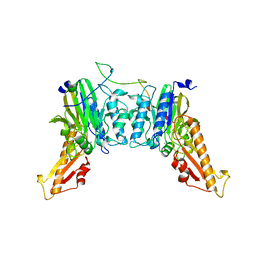 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
4FBW
 
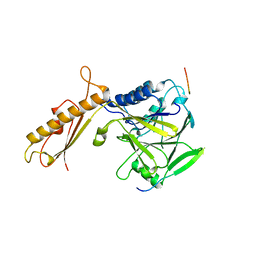 | |
4FCX
 
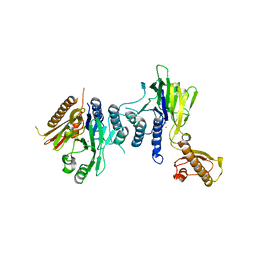 | | S.pombe Mre11 apoenzym | | Descriptor: | DNA repair protein rad32, MANGANESE (II) ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FBK
 
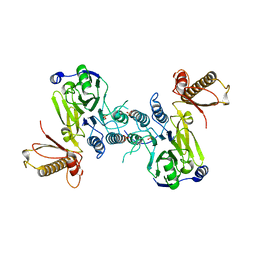 | | Crystal structure of a covalently fused Nbs1-Mre11 complex with one manganese ion per active site | | Descriptor: | DNA repair and telomere maintenance protein nbs1,DNA repair protein rad32 CHIMERIC PROTEIN, MANGANESE (II) ION, SULFATE ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.379 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FBQ
 
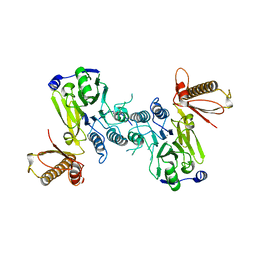 | |
9GMW
 
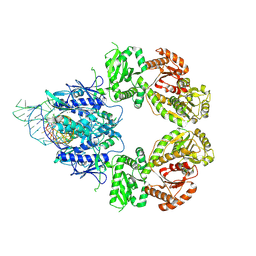 | | SLFN11 WT dimer bound to tRNA-Leu-TAA (pre-cleavage state) | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, RNA (86-MER), ... | | Authors: | Kugler, M, Metzner, F.J, Lammens, K. | | Deposit date: | 2024-08-29 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Phosphorylation-mediated conformational change regulates human SLFN11.
Nat Commun, 15, 2024
|
|
9GMX
 
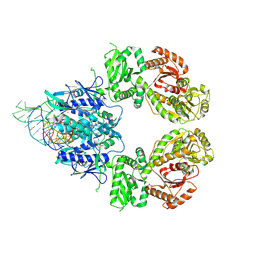 | | SLFN11 WT dimer bound to tRNA-Leu-TAA (post-cleavage state) | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, RNA (5'-R(P*CP*CP*UP*GP*GP*UP*AP*CP*CP*A)-3'), ... | | Authors: | Kugler, M, Metzner, F.J, Lammens, K. | | Deposit date: | 2024-08-29 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Phosphorylation-mediated conformational change regulates human SLFN11.
Nat Commun, 15, 2024
|
|
7Q3Z
 
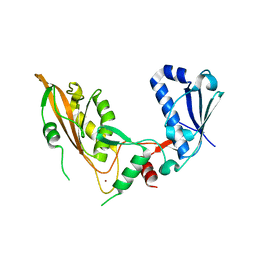 | | DNA/RNA binding protein | | Descriptor: | SODIUM ION, Schlafen family member 5, ZINC ION | | Authors: | Huber, E, Lammens, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 50, 2022
|
|
6S85
 
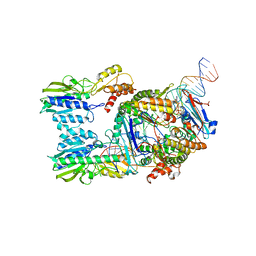 | | Cutting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), DNA (32-MER), ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
5DA9
 
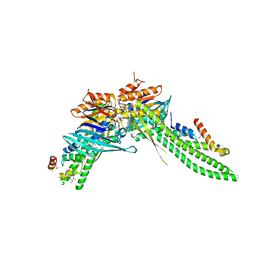 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with the Rad50-binding domain of Mre11 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Putative double-strand break protein, ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
5DAC
 
 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with DNA | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
