7AMA
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | Interleukin-17A, ~{N}-[(2~{S})-1,1-dicyclopropyl-3-[[4-(3,5-dimethyl-1~{H}-pyrazol-4-yl)phenyl]amino]-3-oxidanylidene-propan-2-yl]-2-propan-2-yl-pyrazole-3-carboxamide | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
7AMG
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | (3~{R})-4-[4-[[(2~{S})-2-[[2,2-bis(fluoranyl)-2-phenyl-ethanoyl]amino]-3-(2-chlorophenyl)propanoyl]amino]phenyl]-3-[[(2~{S})-3-methyl-2-[2-[2-[(2-methylpropan-2-yl)oxycarbonylamino]ethoxy]ethanoylamino]butanoyl]amino]butanoic acid, Interleukin-17A | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
6Z1B
 
 | |
1F0J
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 4B2B | | Descriptor: | ARSENIC, MAGNESIUM ION, PHOSPHODIESTERASE 4B, ... | | Authors: | Xu, R.X, Hassell, A.M, Vanderwall, D, Lambert, M.H, Holmes, W.D, Luther, M.A, Rocque, W.J, Milburn, M.V, Zhao, Y, Ke, H, Nolte, R.T. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity.
Science, 288, 2000
|
|
1FM9
 
 | | THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND GI262570 AND CO-ACTIVATOR PEPTIDES. | | Descriptor: | (9cis)-retinoic acid, 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA, ... | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Miller, A.B, Bledsoe, R.K, Milburn, M.V, Kliewer, S.A, Willson, T.M, Xu, H.E. | | Deposit date: | 2000-08-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors.
Mol.Cell, 5, 2000
|
|
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6F4R
 
 | | Human JMJD5 (N308C) in complex with Mn(II), NOG and RCCD1 (139-143) (complex-3) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, JmjC domain-containing protein 5, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
6LLN
 
 | |
6F4O
 
 | | Human JMJD5 in complex with Mn(II) and Succinate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, JmjC domain-containing protein 5, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
6WQD
 
 | | The 1.95 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Kim, Y, Wilamowski, M, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6F4P
 
 | | Human JMJD5 in complex with MN, NOG and RPS6 (129-144) (complex-1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 40S ribosomal protein S6, JmjC domain-containing protein 5, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
6F4Q
 
 | | Human JMJD5 (Q275C) in complex with Mn(II), NOG and RPS6-A138C (129-144) (complex-2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 40S ribosomal protein S6, GLYCEROL, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
7ATL
 
 | | EstCE1, a hydrolase with promiscuous acyltransferase activity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Esterase, ... | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | Discovery and Design of Family VIII Carboxylesterases as Highly Efficient Acyltransferases.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7AH1
 
 | | L19 diabody fragment from immunocytokine L19-IL2 | | Descriptor: | Anti-(ED-B) scFV | | Authors: | Ongaro, T, Guarino, S.R, Scietti, L, Palamini, M, Wulhfard, S, Villa, A, Neri, D, Forneris, F. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inference of molecular structure for characterization and improvement of clinical grade immunocytokines.
J.Struct.Biol., 213, 2021
|
|
7AIC
 
 | | MutS-MutL in clamp state (kinked clamp domain) | | Descriptor: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI7
 
 | | MutS in Intermediate state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*CP*TP*TP*AP*GP*CP*TP*TP*AP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*TP*AP*AP*CP*TP*AP*AP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AIB
 
 | | MutS-MutL in clamp state | | Descriptor: | DNA (30-MER), DNA mismatch repair protein MutL, DNA mismatch repair protein MutS, ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI6
 
 | | MutS in mismatch bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein MutS | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI5
 
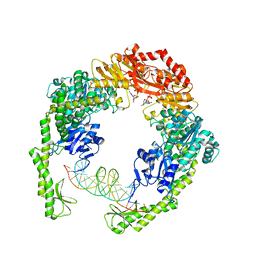 | | MutS in Scanning state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*AP*AP*TP*TP*CP*GP*CP*CP*CP*TP*AP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*TP*AP*GP*GP*GP*CP*GP*AP*AP*TP*TP*GP*GP*GP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6YKG
 
 | |
7BVD
 
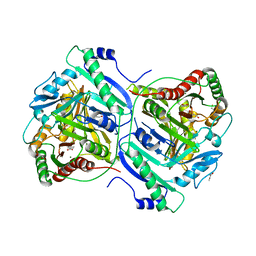 | | Anthranilate synthase component I (TrpE)[Mycolicibacterium smegmatis] | | Descriptor: | Anthranilate synthase component 1, BENZOIC ACID, GLYCEROL, ... | | Authors: | Chen, Y, Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit I of the anthranilate synthase complex of Mycolicibacterium smegmatis
Biochem.Biophys.Res.Commun., 527, 2020
|
|
6Y9K
 
 | |
6YJM
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with the Inhibitor GLPG1972 | | Descriptor: | (5~{S})-5-[3-[(3~{S})-4-[3,5-bis(fluoranyl)phenyl]-3-methyl-piperazin-1-yl]-3-oxidanylidene-propyl]-5-cyclopropyl-imidazolidine-2,4-dione, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Goepfert, A, Leonard, P, Triballeau, N, Fleury, D, Mollat, P, Lamers, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GLPG1972/S201086, a Potent, Selective, and Orally Bioavailable ADAMTS-5 Inhibitor for the Treatment of Osteoarthritis.
J.Med.Chem., 64, 2021
|
|
5LEW
 
 | | DNA polymerase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA polymerase III subunit alpha, SULFATE ION, ... | | Authors: | Banos-Mateos, S, Lang, U.F, Maslen, S.L, Skehel, J.M, Lamers, M.H. | | Deposit date: | 2016-06-30 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-fidelity DNA replication in Mycobacterium tuberculosis relies on a trinuclear zinc center.
Nat Commun, 8, 2017
|
|
5MHP
 
 | | Novel Imidazo[1,2-a]pyridine Derivatives with Potent Autotaxin/ENPP2 Inhibitor Activity | | Descriptor: | 2-[[2-ethyl-8-methyl-6-[4-[2-(3-oxidanylazetidin-1-yl)-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fleury, D, Mueller, I, Lamers, M, Triballeau, N, Mollat, P, Vercheval, L. | | Deposit date: | 2016-11-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of 2-[[2-Ethyl-6-[4-[2-(3-hydroxyazetidin-1-yl)-2-oxoethyl]piperazin-1-yl]-8-methylimidazo[1,2-a]pyridin-3-yl]methylamino]-4-(4-fluorophenyl)thiazole-5-carbonitrile (GLPG1690), a First-in-Class Autotaxin Inhibitor Undergoing Clinical Evaluation for the Treatment of Idiopathic Pulmonary Fibrosis.
J. Med. Chem., 60, 2017
|
|
