8BYA
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
8BYL
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex | | Descriptor: | Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, S-phase kinase-associated protein 1, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
8BZO
 
 | | Cryo-EM structure of CDK2-CyclinA in complex with p27 from the SCFSKP2 E3 ligase Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-15 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
3AKA
 
 | | Structural basis for prokaryotic calcium-mediated regulation by a Streptomyces coelicolor calcium-binding protein | | Descriptor: | CALCIUM ION, Putative calcium binding protein | | Authors: | Zhao, X, Pang, H, Wang, S, Zhou, W, Yang, K, Bartlam, M. | | Deposit date: | 2010-07-09 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prokaryotic calciummediated regulation by a Streptomyces coelicolor calcium binding protein
Protein Cell, 1, 2010
|
|
6F4M
 
 | |
6F4T
 
 | | Human JMJD5 (W414C) in complex with Mn(II), NOG and RCCD1 (139-143) (complex-5) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, JmjC domain-containing protein 5, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
6F4N
 
 | |
6F4S
 
 | | Human JMJD5 (N308C) in complex with Mn(II), 2OG and RCCD1 (139-143) (complex-4) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-OXOGLUTARIC ACID, GLYCEROL, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
1U9E
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-397 | | Descriptor: | 2-(4-HYDROXY-PHENYL)BENZOFURAN-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-09 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design Of Estrogen Receptor-beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
3O9B
 
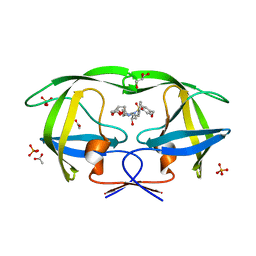 | | Crystal Structure of wild-type HIV-1 Protease in Complex with kd25 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, ACETATE ION, GLYCEROL, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3O9C
 
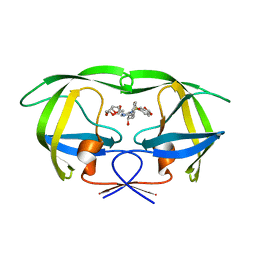 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd20 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzodioxol-5-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Pol polyprotein | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
1K7L
 
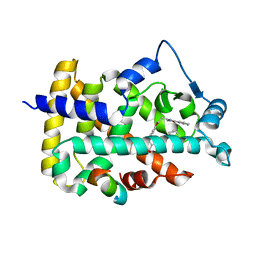 | | The 2.5 Angstrom resolution crystal structure of the human PPARalpha ligand binding domain bound with GW409544 and a co-activator peptide. | | Descriptor: | 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor alpha, YTTRIUM (III) ION, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KKQ
 
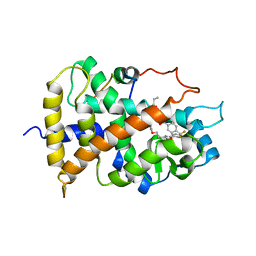 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
1UDE
 
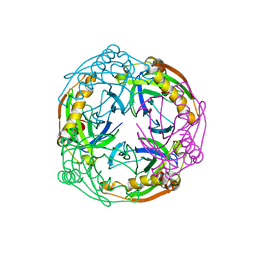 | | Crystal structure of the Inorganic pyrophosphatase from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Liu, B, Gao, R, Zhou, W, Bartlam, M, Rao, Z. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of the hyperthermophilic inorganic pyrophosphatase from the archaeon Pyrococcus horikoshii.
Biophys.J., 86, 2004
|
|
1K74
 
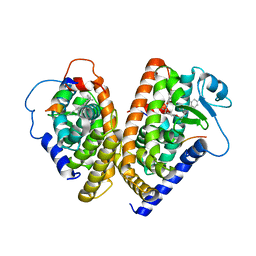 | | The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides. | | Descriptor: | (9cis)-retinoic acid, 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor gamma, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3NV6
 
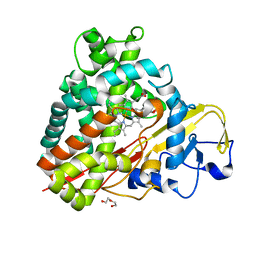 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
4X42
 
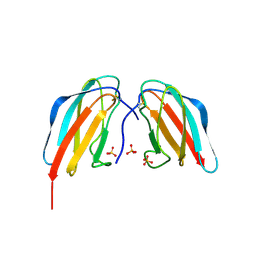 | | Crystal structure of DEN4 ED3 mutant with epitope two residues substituted from DEN3 ED3 | | Descriptor: | Envelope protein E, SULFATE ION | | Authors: | Kulkarni, M.R, Islam, M.M, Numoto, N, Elahi, M.M, Ito, N, Kuroda, Y. | | Deposit date: | 2014-12-02 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and biophysical analysis of sero-specific immune responses using epitope grafted Dengue ED3 mutants.
Biochim.Biophys.Acta, 1854, 2015
|
|
4WP6
 
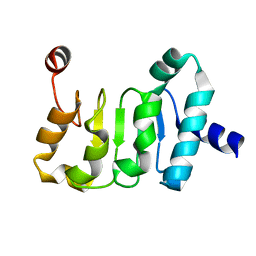 | | Structure of the Mex67 LRR domain from Chaetomium thermophilum | | Descriptor: | mRNA export protein | | Authors: | Aibara, S, Valkov, E, Lamers, M, Stewart, M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of the principal mRNA-export factor Mex67-Mtr2 from Chaetomium thermophilum.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7PKR
 
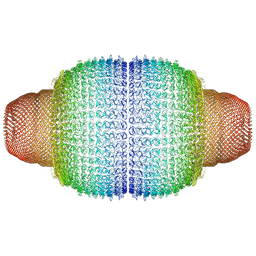 | | Vault structure in primmed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7PKZ
 
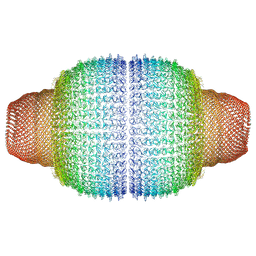 | | Vault structure in committed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7PKY
 
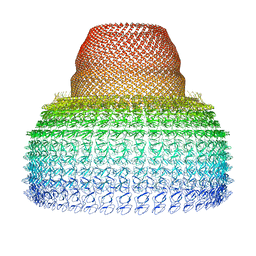 | | Half-vault structure | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7L5A
 
 | | Crystal structure of the photosensory module from Xanthomonas campestris bacteriophytochrome XccBphP in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7L59
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP-G454E variant from Xanthomonas campestris in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7AC0
 
 | | Epoxide hydrolase CorEH without ligand | | Descriptor: | Soluble epoxide hydrolase | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Promiscuous Dehalogenase Activity of the Epoxide Hydrolase CorEH from Corynebacterium sp. C12
Acs Catalysis, 11, 2021
|
|
