4JXM
 
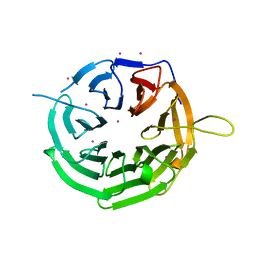 | | Crystal structure of RRP9 WD40 repeats | | Descriptor: | U3 small nucleolar RNA-interacting protein 2, UNKNOWN ATOM OR ION | | Authors: | Wu, X, Tempel, W, Xu, C, El Bakkouri, M, He, H, Seitova, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of RRP9 WD40 repeats
To be Published
|
|
1SMF
 
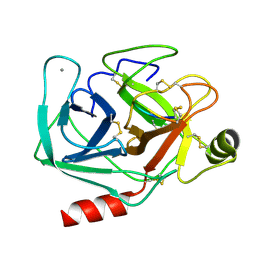 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | Authors: | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | Deposit date: | 1992-10-24 | | Release date: | 1994-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
8ITE
 
 | |
8V04
 
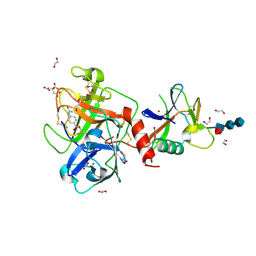 | | High resolution TMPRSS2 structure following acylation by nafamostat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Levon, H, Arrowsmith, C. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High resolution TMPRSS2 structure following acylation by nafamostat
To Be Published
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
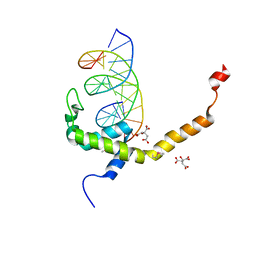 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7DTQ
 
 | |
2PW6
 
 | | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12 | | Descriptor: | Uncharacterized protein ygiD, ZINC ION | | Authors: | Newton, M.G, Takagi, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokayama, S, Li, Y, Chen, L, Zhu, J, Ruble, J, Liu, Z.J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12.
To be Published
|
|
2AI3
 
 | | Purine nucleoside phosphorylase from calf spleen | | Descriptor: | (2S,4R,6R,6AS)-4-(2-AMINO-6-OXO-1,6-DIHYDROPURIN-9-YL)-6-(HYDROXYMETHYL)-TETRAHYDROFURO[3,4-D][1,3]DIOXOL-2-YLPHOSPHONI C ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | Deposit date: | 2005-07-28 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1APH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 7), INSULIN B CHAIN (PH 7) | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
4KY1
 
 | | humanized HP1/2 Fab | | Descriptor: | IMMUNOGLOBULIN IGG1 FAB, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Arndt, J.W, Hanf, K.J.M, Lugovskoy, A, Chen, L.L, Jarpe, M, Boriack-Sjodin, A, Li, Y, van Vlijmen, H, Pepinsky, B, Taylor, F, Silvian, L, Taveras, A. | | Deposit date: | 2013-05-28 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Antibody humanization by redesign of complementarity-determining region residues proximate to the acceptor framework.
METHODS (SAN DIEGO), 65, 2014
|
|
3MD1
 
 | | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Li, H, Shi, H, Li, Y, Cui, Y, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1)
To be published
|
|
1JHN
 
 | | Crystal Structure of the Lumenal Domain of Calnexin | | Descriptor: | CALCIUM ION, calnexin | | Authors: | Schrag, J.D, Bergeron, J.M, Li, Y, Borisova, S, Hahn, M, Thomas, D.Y, Cygler, M. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of calnexin, an ER chaperone involved in quality control of protein folding.
Mol.Cell, 8, 2001
|
|
3FCX
 
 | | Crystal structure of human esterase D | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-formylglutathione hydrolase | | Authors: | Wu, D, Li, Y, Song, G, Zhang, D, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-11-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human esterase D: a potential genetic marker of retinoblastoma
Faseb J., 23, 2009
|
|
3ZDM
 
 | | Crystal structure of the Sgt2 N domain and the Get5 UBL domain complex | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT- CONTAINING PROTEIN 2, UBIQUITIN-LIKE PROTEIN MDY2 | | Authors: | Tung, J.-Y, Li, Y.-C, Hsiao, C.-D. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure of the Sgt2 Dimerization Domain Complexed with the Get5 Ubl Domain Involved in the Targeting of Tail-Anchored Membrane Proteins to the Endoplasmic Reticulum.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I52
 
 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
5XI2
 
 | | BRD4 bound with compound Bdi2 | | Descriptor: | (3~{R})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{S})-1-(4-methylphenyl)ethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2017-04-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | BRD4 bound with compound Bdi2
To Be Published
|
|
6LX3
 
 | | Cryo-EM structure of human secretory immunoglobulin A | | Descriptor: | Immunoglobulin J chain, Interleukin-2,Immunoglobulin heavy constant alpha 1, Polymeric immunoglobulin receptor | | Authors: | Wang, Y, Wang, G, Li, Y, Xiao, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin.
Cell Res., 30, 2020
|
|
4QN1
 
 | | Crystal Structure of a Functionally Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Walker, J.R, Guan, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a Function Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH
To be Published
|
|
4NQJ
 
 | | Structure of coiled-coil domain | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, E3 ubiquitin-protein ligase TRIM69 | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2013-11-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural insights into the TRIM family of ubiquitin E3 ligases.
Cell Res., 24, 2014
|
|
8H1C
 
 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (one in tRNA binding state and the other in tRNA locked state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) (74-MER) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-10-02 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
3OMU
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of a thienopyrimidine derivative | | Descriptor: | 2-amino-4-{2,4-dichloro-5-[2-(diethylamino)ethoxy]phenyl}-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, Heat shock protein 83 | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Cossar, D, Li, Y, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-27 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
8K8S
 
 | | F8-A22-E4 complex of MPXV in complex with DNA and Ara-CTP | | Descriptor: | 4-amino-1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, DNA (5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*TP*AP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Shen, Y.P, Li, Y.N, Yan, R.H. | | Deposit date: | 2023-07-31 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for the inhibition mechanism of the DNA polymerase holoenzyme from mpox virus.
Structure, 32, 2024
|
|
