7ZJE
 
 | | C16-2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2,Enhanced green fluorescent protein | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-10 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
6H0K
 
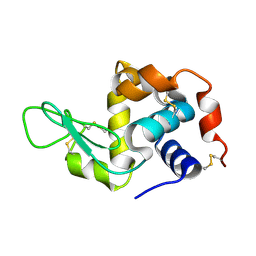 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, the first MHz free electron laser, 7.47 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0L
 
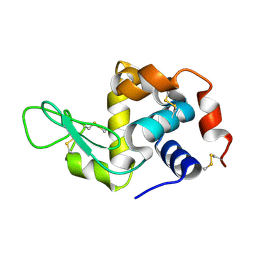 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, 9.22 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
7ZJG
 
 | | Probenecid | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
7ZJI
 
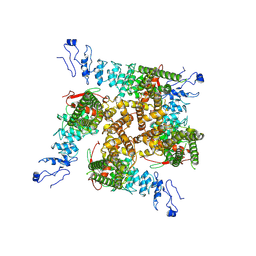 | |
1S1G
 
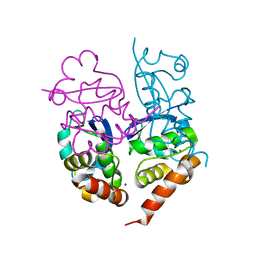 | | Crystal Structure of Kv4.3 T1 Domain | | Descriptor: | Potassium voltage-gated channel subfamily D member 3, ZINC ION | | Authors: | Scannevin, R.H, Wang, K.W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.X, Xu, Z.B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
7ZJH
 
 | | TRPV2-C16+Pro-2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
7ZJD
 
 | |
1S36
 
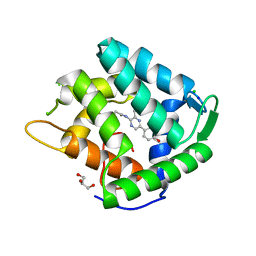 | | Crystal structure of a Ca2+-discharged photoprotein: Implications for the mechanisms of the calcium trigger and the bioluminescence | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, ... | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.-J, Lee, J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-01-12 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a Ca2+-discharged photoprotein: implications for mechanisms of the calcium trigger and bioluminescence
J.Biol.Chem., 279, 2004
|
|
4ZZO
 
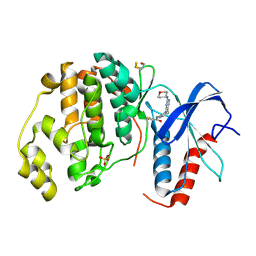 | | Human ERK2 in complex with an irreversible inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 1, N-[2-[[5-chloranyl-2-(oxan-4-ylamino)pyrimidin-4-yl]amino]phenyl]propanamide, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
1RYI
 
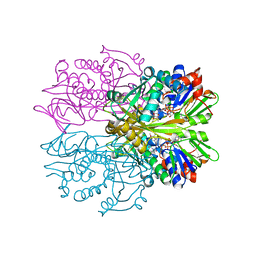 | | STRUCTURE OF GLYCINE OXIDASE WITH BOUND INHIBITOR GLYCOLATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE OXIDASE, GLYCOLIC ACID | | Authors: | Moertl, M, Diederichs, K, Welte, W, Pollegioni, L, Molla, G, Motteran, L, Andriolo, G, Pilone, M.S. | | Deposit date: | 2003-12-22 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function correlation in glycine oxidase from Bacillus subtilis
J.Biol.Chem., 279, 2004
|
|
5MMN
 
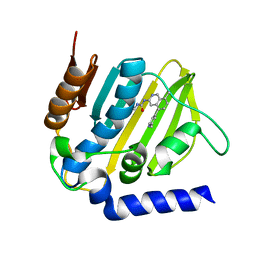 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[8-methyl-5-(2-methylpyridin-4-yl)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
6ENL
 
 | |
1SBK
 
 | | X-RAY STRUCTURE OF YDII_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER29. | | Descriptor: | Hypothetical protein ydiI, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Vorobiev, S.M, Lee, I, Forouhar, F, Ma, L, Chiang, Y, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Structure of YDII_ECOLI Northeast Structural Genomics Consortium Target ER29
To be Published
|
|
1NYX
 
 | | Ligand binding domain of the human peroxisome proliferator activated receptor gamma in complex with an agonist | | Descriptor: | (2S)-2-ETHOXY-3-{4-[2-(10H-PHENOXAZIN-10-YL)ETHOXY]PHENYL}PROPANOIC ACID, peroxisome proliferator activated receptor gamma | | Authors: | Ebdrup, S, Pettersson, I, Rasmussen, H.B, Deussen, H.-J, Frost Jensen, A, Mortensen, S.B, Fleckner, J, Pridal, L, Nygaard, L, Sauerberg, P. | | Deposit date: | 2003-02-14 | | Release date: | 2003-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and biological and structural characterization of the dual-acting peroxisome proliferator-activated receptor alpha/gamma agonist ragaglitazar
J.MED.CHEM., 46, 2003
|
|
1G9F
 
 | | CRYSTAL STRUCTURE OF THE SOYBEAN AGGLUTININ IN A COMPLEX WITH A BIANTENNARY BLOOD GROUP ANTIGEN ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Buts, L, Hamelryck, T.W, Dao-Thi, M.-H, Loris, R, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-23 | | Release date: | 2001-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
5LV5
 
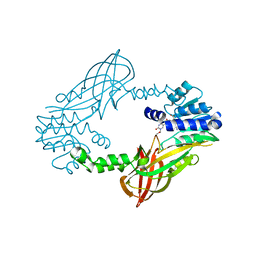 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1458 | | Descriptor: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]-[(3~{S})-3-azanyl-4-oxidanyl-4-oxidanylidene-butyl]azanium, Protein arginine N-methyltransferase 6 | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
6OV8
 
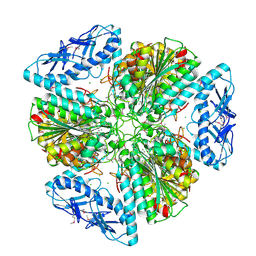 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
4OHE
 
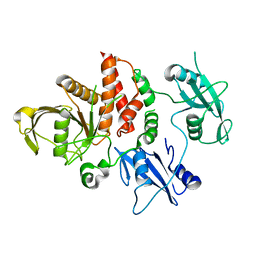 | | LEOPARD Syndrome-Associated SHP2/G464A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
5CUH
 
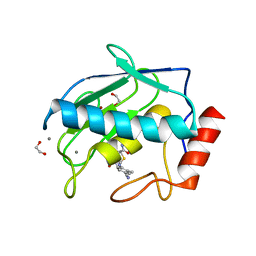 | | Crystal structure MMP-9 complexes with a constrained hydroxamate based inhibitor LT4 | | Descriptor: | (4S)-3-{[4-(4-cyano-2-methylphenyl)piperazin-1-yl]sulfonyl}-N-hydroxy-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Tepshi, L, Vera, L, Nuti, E, Rosalia, L, Rossello, A, Stura, E.A. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a new selective inhibitor of A Disintegrin And Metalloprotease 10 (ADAM-10) able to reduce the shedding of NKG2D ligands in Hodgkin's lymphoma cell models.
Eur.J.Med.Chem., 111, 2016
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
1UNL
 
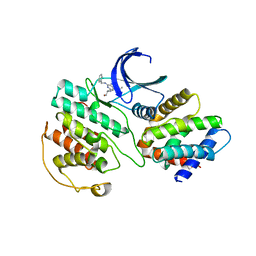 | | Structural mechanism for the inhibition of CD5-p25 from the roscovitine, aloisine and indirubin. | | Descriptor: | CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1, R-ROSCOVITINE | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UYK
 
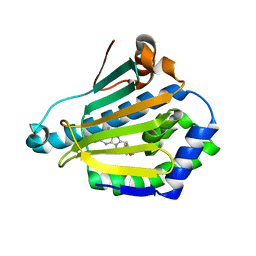 | | Human Hsp90-alpha with 8-Benzo[1,3]dioxol-,5-ylmethyl-9-butyl-2-fluoro-9H-purin-6-ylamine | | Descriptor: | 8-BENZO[1,3]DIOXOL-,5-YLMETHYL-9-BUTYL-2-FLUORO-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
5DG5
 
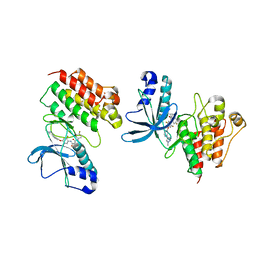 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET IN COMPLEX WITH ALTIRATINIB ANALOG DP-4157 | | Descriptor: | Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[2-(1-methyl-1H-pyrazol-4-yl)pyridin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxam ide | | Authors: | Smith, B.D, Kaufman, M.D, Leary, C.B, Turner, B.A, Wise, S.A, Ahn, Y.M, Booth, R.J, Caldwell, T.M, Ensinger, C.L, Hood, M.M, Lu, W.-P, Patt, T.W, Patt, W.C, Rutkoski, T.J, Samarakoon, T, Telikepalli, H, Vogeti, L, Vogeti, S, Yates, K.M, Chun, L, Stewart, L.J, Clare, M, Flynn, D.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2.
Mol.Cancer Ther., 14, 2015
|
|
8RGH
 
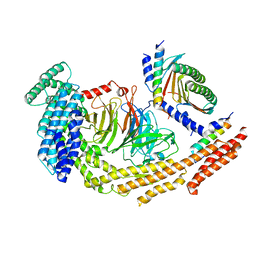 | | Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
