3KEX
 
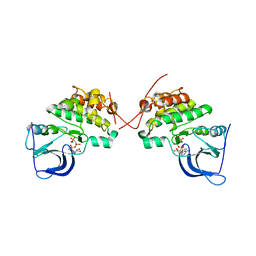 | | Crystal structure of the catalytically inactive kinase domain of the human epidermal growth factor receptor 3 (HER3) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Jura, N, Shan, Y, Cao, X, Shaw, D.E, Kuriyan, J. | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2POL
 
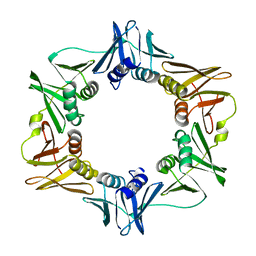 | |
1IAS
 
 | | CYTOPLASMIC DOMAIN OF UNPHOSPHORYLATED TYPE I TGF-BETA RECEPTOR CRYSTALLIZED WITHOUT FKBP12 | | Descriptor: | SULFATE ION, TGF-BETA RECEPTOR TYPE I | | Authors: | Huse, M, Muir, T.W, Chen, Y.-G, Kuriyan, J, Massague, J. | | Deposit date: | 2001-03-23 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The TGF beta receptor activation process: an inhibitor- to substrate-binding switch.
Mol.Cell, 8, 2001
|
|
1OPJ
 
 | | Structural basis for the auto-inhibition of c-Abl tyrosine kinase | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Young, M.A, Scheffzek, K, Veach, D, Bornmann, W, Clarkson, B, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the autoinhibition of c-Abl tyrosine kinase
Cell(Cambridge,Mass.), 112, 2003
|
|
1FPU
 
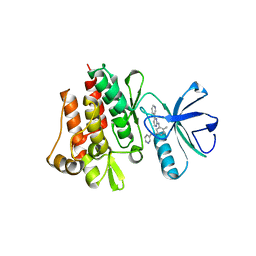 | | CRYSTAL STRUCTURE OF ABL KINASE DOMAIN IN COMPLEX WITH A SMALL MOLECULE INHIBITOR | | Descriptor: | N-[4-METHYL-3-[[4-(3-PYRIDINYL)-2-PYRIMIDINYL]AMINO]PHENYL]-3-PYRIDINECARBOXAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL | | Authors: | Schindler, T, Bornmann, W, Pellicena, P, Miller, W.T, Clarkson, B, Kuriyan, J. | | Deposit date: | 2000-08-31 | | Release date: | 2000-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism for STI-571 inhibition of abelson tyrosine kinase.
Science, 289, 2000
|
|
2HNH
 
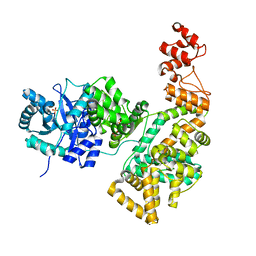 | | Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III | | Descriptor: | DNA polymerase III alpha subunit, PHOSPHATE ION | | Authors: | Meindert, M.H, Georgescu, R.E, Lee, S, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HQA
 
 | | Crystal structure of the catalytic alpha subunit of E. Coli replicative DNA polymerase III | | Descriptor: | DNA polymerase III alpha subunit, PHOSPHATE ION | | Authors: | Lamers, M.H, Georgescu, R.E, Lee, S.G, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
3U60
 
 | | Structure of T4 Bacteriophage Clamp Loader Bound To Open Clamp, DNA and ATP Analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
3U5Z
 
 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
3F6X
 
 | | c-Src kinase domain in complex with small molecule inhibitor | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, [4-({4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl}amino)phenyl]acetonitrile | | Authors: | Seeliger, M.A, Statsuk, A.V, Maly, D.J, Patrick, P.Z, Kuriyan, J, Shokat, K.M. | | Deposit date: | 2008-11-06 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tuning a three-component reaction for trapping kinase substrate complexes.
J.Am.Chem.Soc., 130, 2008
|
|
3K4X
 
 | | Eukaryotic Sliding Clamp PCNA Bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), Proliferating cell nuclear antigen | | Authors: | McNally, R, Kuriyan, J. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Analysis of the role of PCNA-DNA contacts during clamp loading.
Bmc Struct.Biol., 10, 2010
|
|
1JQL
 
 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of beta-delta (1-140) | | Descriptor: | DNA Polymerase III, BETA CHAIN, DELTA SUBUNIT | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
3GEQ
 
 | | Structural basis for the chemical rescue of Src kinase activity | | Descriptor: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Muratore, K.E, Seeliger, M.A, Wang, Z, Fomina, D, Neiswinger, J, Havranek, J.J, Baker, D, Kuriyan, J, Cole, P.A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative analysis of mutant tyrosine kinase chemical rescue.
Biochemistry, 48, 2009
|
|
3GLF
 
 | |
3GLG
 
 | | Crystal Structure of a Mutant (gammaT157A) E. coli Clamp Loader Bound to Primer-Template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Simonetta, K.R, Seyedin, S.N, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GLI
 
 | | Crystal Structure of the E. coli clamp loader bound to Primer-Template DNA and Psi Peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Simonetta, K.R, Cantor, A.J, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
1JQJ
 
 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of the beta-delta complex | | Descriptor: | DNA polymerase III, beta chain, delta subunit | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
2IJE
 
 | |
3ET6
 
 | | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase | | Descriptor: | PHOSPHATE ION, Soluble guanylyl cyclase beta | | Authors: | Winger, J.A, Derbyshire, E.R, Lamers, M.H, Marletta, M.A, Kuriyan, J. | | Deposit date: | 2008-10-07 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase.
Bmc Struct.Biol., 8, 2008
|
|
1HKX
 
 | | Crystal structure of calcium/calmodulin-dependent protein kinase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II ALPHA CHAIN, CHLORIDE ION, ... | | Authors: | Hoelz, A, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2003-03-12 | | Release date: | 2003-06-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of a Tetradecameric Assembly of the Association Domain of Ca2+/Calmodulin-Dependent Kinase II
Mol.Cell, 11, 2003
|
|
2II0
 
 | | Crystal Structure of catalytic domain of Son of sevenless (Rem-Cdc25) in the absence of Ras | | Descriptor: | Son of sevenless homolog 1 | | Authors: | Freedman, T.S, Sondermann, H, Friedland, G.D, Kortemme, T, Bar-Sagi, D, Marqusee, S, Kuriyan, J. | | Deposit date: | 2006-09-27 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Ras-induced conformational switch in the Ras activator Son of sevenless.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3SOA
 
 | | Full-length human CaMKII | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Calcium/calmodulin-dependent protein kinase type II subunit alpha with a beta 7 linker | | Authors: | Chao, L.H, Kuriyan, J. | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5501 Å) | | Cite: | A Mechanism for Tunable Autoinhibition in the Structure of a Human Ca(2+)/Calmodulin- Dependent Kinase II Holoenzyme.
Cell(Cambridge,Mass.), 146, 2011
|
|
2HCK
 
 | | SRC FAMILY KINASE HCK-QUERCETIN COMPLEX | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, CALCIUM ION, HEMATOPOETIC CELL KINASE HCK | | Authors: | Sicheri, F, Moarefi, I, Kuriyan, J. | | Deposit date: | 1997-02-25 | | Release date: | 1997-08-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Src family tyrosine kinase Hck.
Nature, 385, 1997
|
|
1OPK
 
 | | Structural basis for the auto-inhibition of c-Abl tyrosine kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Young, M.A, Scheffzek, K, Veach, D, Bornmann, W, Clarkson, B, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the autoinhibition of c-Abl tyrosine kinase
Cell(Cambridge,Mass.), 112, 2003
|
|
3U61
 
 | | Structure of T4 Bacteriophage Clamp Loader Bound To Closed Clamp, DNA and ATP Analog and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
