4R9U
 
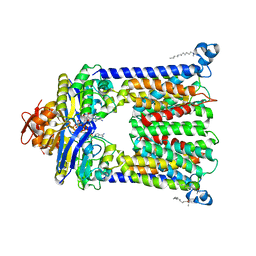 | | Structure of vitamin B12 transporter BtuCD in a nucleotide-bound outward facing state | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Korkhov, V.M, Mireku, S.A, Veprintsev, D.B, Locher, K.P. | | Deposit date: | 2014-09-08 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4FI3
 
 | | Structure of vitamin B12 transporter BtuCD-F in a nucleotide-bound state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Vitamin B12 import ATP-binding protein BtuD, ... | | Authors: | Korkhov, V.M, Mireku, S.A, Locher, K.P. | | Deposit date: | 2012-06-07 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.466 Å) | | Cite: | Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F.
Nature, 490, 2012
|
|
4DBL
 
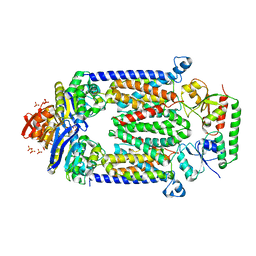 | | Crystal structure of E159Q mutant of BtuCDF | | Descriptor: | PHOSPHATE ION, SULFATE ION, Vitamin B12 import ATP-binding protein BtuD, ... | | Authors: | Korkhov, V.M, Mireku, S.M, Hvorup, R.N, Locher, K.P. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Asymmetric states of vitamin B12 transporter BtuCD are not discriminated by its cognate substrate binding protein BtuF.
Febs Lett., 586, 2012
|
|
7NUR
 
 | |
6R4P
 
 | |
6RMG
 
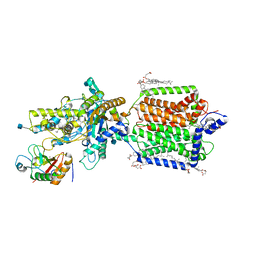 | | Structure of PTCH1 bound to a modified Hedgehog ligand ShhN-C24II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Korkhov, V.M, Qi, C. | | Deposit date: | 2019-05-06 | | Release date: | 2019-10-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of sterol recognition by human hedgehog receptor PTCH1.
Sci Adv, 5, 2019
|
|
6R3Q
 
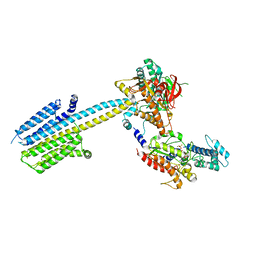 | |
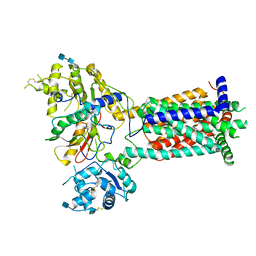
6TD6
 
 | |
6TBU
 
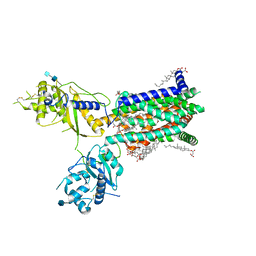 | | Structure of Drosophila melanogaster Dispatched | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Korkhov, V.M, Cannac, F. | | Deposit date: | 2019-11-04 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the Hedgehog release protein Dispatched.
Sci Adv, 6, 2020
|
|
8BUZ
 
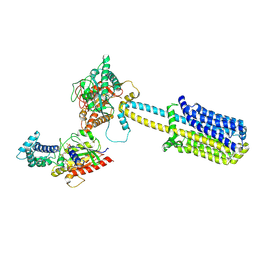 | | Structure of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase type 8, FORSKOLIN, ... | | Authors: | Khanppnavar, B, Korkhov, V.M, Mehta, V. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Regulatory sites of CaM-sensitive adenylyl cyclase AC8 revealed by cryo-EM and structural proteomics.
Embo Rep., 25, 2024
|
|
8BV5
 
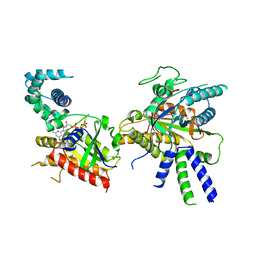 | | Focus refinement of soluble domain of Adenylyl cyclase 8 bound to stimulatory G protein, Forskolin, ATPalphaS, and Ca2+/Calmodulin in lipid nanodisc conditions | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase type 8, FORSKOLIN, ... | | Authors: | Khanppnavar, B, Korkhov, V.M. | | Deposit date: | 2023-01-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Regulatory sites of CaM-sensitive adenylyl cyclase AC8 revealed by cryo-EM and structural proteomics.
Embo Rep., 25, 2024
|
|
8QOJ
 
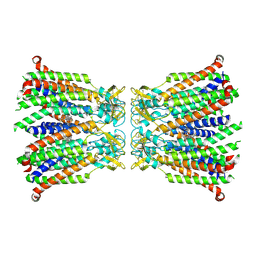 | |
8QJH
 
 | |
7YZ9
 
 | | Structure of catalytic domain of Rv1625c bound to nanobody NB4 | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, GLYCEROL, ... | | Authors: | Khanppnavar, B, Mehta, V.J, Iype, T, Korkhov, V.M. | | Deposit date: | 2022-02-19 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases.
Elife, 11, 2022
|
|
7YZK
 
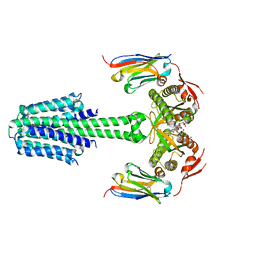 | | Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, MANGANESE (II) ION, ... | | Authors: | Mehta, V, Khanppnavar, B, Korkhov, V.M. | | Deposit date: | 2022-02-20 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases.
Elife, 11, 2022
|
|
7YZI
 
 | | Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, MANGANESE (II) ION, ... | | Authors: | Mehta, V, Khanppnavar, B, Korkhov, V.M. | | Deposit date: | 2022-02-20 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases.
Elife, 11, 2022
|
|
8JPS
 
 | | Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map) | | Descriptor: | Atypical chemokine receptor 1, C-C motif chemokine 7 | | Authors: | Banerjee, R, Khanppnavar, B, Maharana, J, Saha, S, Korkhov, V.M, Shukla, A.K. | | Deposit date: | 2023-06-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular mechanism of distinct chemokine engagement and functional divergence of the human Duffy antigen receptor.
Cell, 187, 2024
|
|
8QJF
 
 | |
8QKI
 
 | |
8QK6
 
 | |
8QKO
 
 | |
8R7R
 
 | |
5O5K
 
 | | X-ray structure of a bacterial adenylyl cyclase soluble domain | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, MANGANESE (II) ION, ... | | Authors: | Vercellino, I, Korkhov, V.M. | | Deposit date: | 2017-06-02 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Role of the nucleotidyl cyclase helical domain in catalytically active dimer formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5M2Q
 
 | | Structure of cobinamide-bound BtuF mutant W66F, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-13 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
5O5L
 
 | |
