8BDU
 
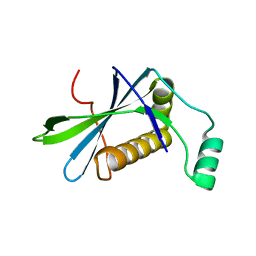 | | H33 variant of DoBi scaffold based on PIH1D1 N-terminal domain | | Descriptor: | PIH1 domain-containing protein 1 | | Authors: | Kolenko, P, Mikulecky, P, Pham, P.N, Schneider, B. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Diffraction anisotropy and paired refinement: crystal structure of H33, a protein binder to interleukin 10.
J.Appl.Crystallogr., 56, 2023
|
|
3HUP
 
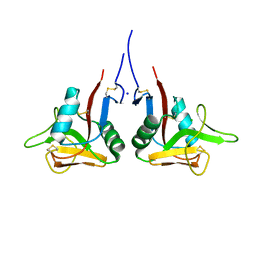 | | High-resolution structure of the extracellular domain of human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69, SODIUM ION | | Authors: | Kolenko, P, Dohnalek, J, Skalova, T, Hasek, J, Duskova, J, Vanek, O, Bezouska, K. | | Deposit date: | 2009-06-15 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | The high-resolution structure of the extracellular domain of human CD69 using a novel polymer
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2RGS
 
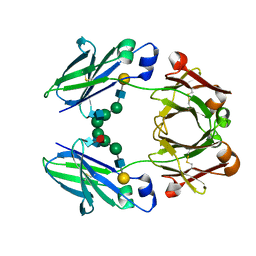 | | FC-fragment of monoclonal antibody IGG2B from Mus musculus | | Descriptor: | Ig gamma-2B heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J, Hasek, J. | | Deposit date: | 2007-10-05 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | New insights into intra- and intermolecular interactions of immunoglobulins: crystal structure of mouse IgG2b-Fc at 2.1-A resolution
Immunology, 126, 2008
|
|
6F1E
 
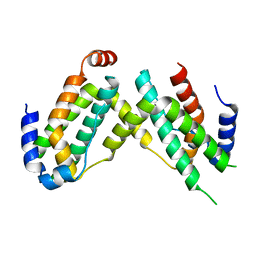 | | Crystal structure of olive flounder [Paralichthys olivaceus] interferon gamma at 2.3 Angstrom resolution | | Descriptor: | Interferon gamma | | Authors: | Kolenko, P, Kolarova, L, Zahradnik, J, Schneider, B. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Interferons type II and their receptors R1 and R2 in fish species: Evolution, structure, and function.
Fish Shellfish Immunol., 79, 2018
|
|
6GG1
 
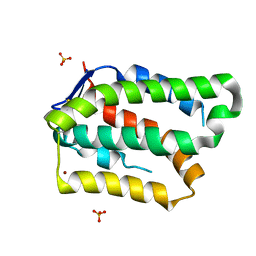 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
3M9Z
 
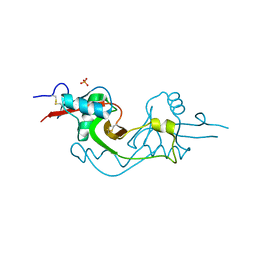 | | Crystal Structure of extracellular domain of mouse NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular architecture of mouse activating NKR-P1 receptors.
J.Struct.Biol., 175, 2011
|
|
7AVC
 
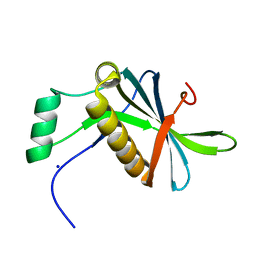 | | DoBi scaffold based on PIH1D1 N-terminal domain | | Descriptor: | GLYCEROL, PIH1 domain-containing protein 1, SODIUM ION | | Authors: | Kolenko, P, Pham, N.P, Pavlicek, J, Mikulecky, P, Schneider, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein Binder (ProBi) as a New Class of Structurally Robust Non-Antibody Protein Scaffold for Directed Evolution.
Viruses, 13, 2021
|
|
5EH1
 
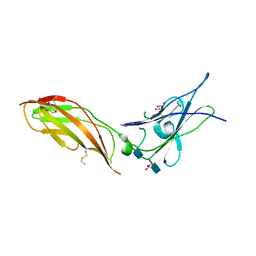 | | Crystal structure of the extracellular part of receptor 2 of human interferon gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, GLYCEROL, ... | | Authors: | Kolenko, P, Mikulecky, P, Zahradnik, J, Dohnalek, J, Koval, T, Cerny, J, Necasova, I, Schneider, B. | | Deposit date: | 2015-10-27 | | Release date: | 2016-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human interferon-gamma receptor 2 reveals the structural basis for receptor specificity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EPD
 
 | | Crystal structure of Glycerol Trinitrate Reductase XdpB from Agrobacterium sp. R89-1 (Apo form) | | Descriptor: | Glycerol trinitrate reductase | | Authors: | Kolenko, P, Zahradnik, J, Zuskova, I, Cerny, J, Palyzova, A, Kyslikova, E, Schneider, B. | | Deposit date: | 2015-11-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of XdpB, the bacterial old yellow enzyme, in an FMN-free form.
PLoS ONE, 13, 2018
|
|
6ROR
 
 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from C. hominis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ROS
 
 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from C. hominis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ROU
 
 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from H. parasuis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4FWU
 
 | | Crystal structure of glutaminyl cyclase from drosophila melanogaster in space group I4 | | Descriptor: | 1,2-ETHANEDIOL, CG32412, SULFATE ION, ... | | Authors: | Kolenko, P, Koch, B, Stubbs, M.T. | | Deposit date: | 2012-07-02 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of glutaminyl cyclase from Drosophila melanogaster in space group I4.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4F9U
 
 | | Structure of glycosylated glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CG32412, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4F9V
 
 | | Structure of C113A/C136A mutant variant of glycosylated glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CG32412, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4FBE
 
 | | Crystal structure of the C136A/C164A variant of mitochondrial isoform of glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CG5976, isoform B, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4FAI
 
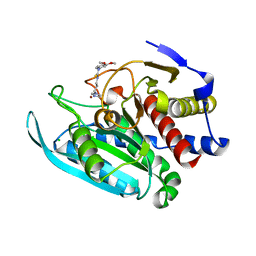 | | Crystal structure of mitochondrial isoform of glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CG5976, isoform B, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
3T3A
 
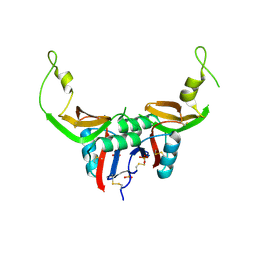 | | Crystal structure of H107R mutant of extracellular domain of mouse receptor NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the H107R variant of the extracellular domain of mouse NKR-P1A at 2.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
7QTB
 
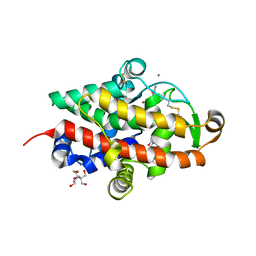 | | S1 nuclease from Aspergillus oryzae in complex with cytidine-5'-monophosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QTA
 
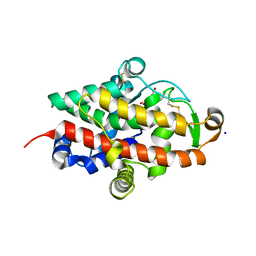 | | S1 nuclease from Aspergillus oryzae in complex with uridine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6OTD
 
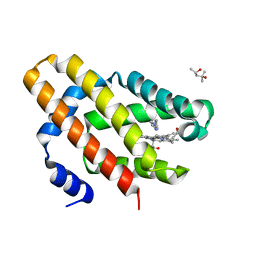 | | Globin sensor domain of AfGcHK in monomeric form, with imidazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Globin-coupled histidine kinase, IMIDAZOLE, ... | | Authors: | Skalova, T, Dohnalek, J, Kolenko, P, Stranava, M, Lengalova, A, Martinkova, M. | | Deposit date: | 2019-05-03 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Disruption of the dimerization interface of the sensing domain in the dimeric heme-based oxygen sensorAfGcHK abolishes bacterial signal transduction.
J.Biol.Chem., 295, 2020
|
|
3CG8
 
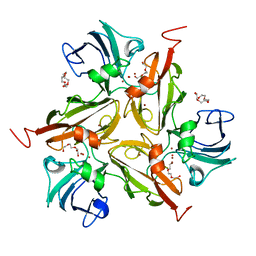 | | Laccase from Streptomyces coelicolor | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, TETRAETHYLENE GLYCOL, ... | | Authors: | Skalova, T, Dohnalek, J, Ostergaard, L.H, Ostergaard, P.R, Kolenko, P, Duskova, J, Hasek, J. | | Deposit date: | 2008-03-05 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | The Structure of the Small Laccase from Streptomyces coelicolor Reveals a Link between Laccases and Nitrite Reductases.
J.Mol.Biol., 385, 2009
|
|
6ZE7
 
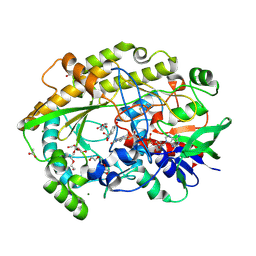 | | Chaetomium thermophilum FAD-dependent oxidoreductase in complex with 4-nitrophenol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZE5
 
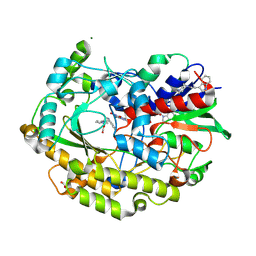 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZE6
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 4-nitrocatechol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITROCATECHOL, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
