6HSX
 
 | | Thrombin in Complex with a D-Phe-Pro-diaminopyridine derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[[2,6-bis(azanyl)pyridin-4-yl]methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2018-10-02 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
6I0W
 
 | | Human Carbonic Anhydrase II in complex with 4-Methoxybenzenesulfonamide | | Descriptor: | 4-methoxybenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-10-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6HR3
 
 | | Human Carbonic Anhydrase II in complex with 4-Propylbenzenesulfonamide | | Descriptor: | 4-propylbenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6HXD
 
 | | Human Carbonic Anhydrase II in complex with 4-Butylbenzenesulfonamide | | Descriptor: | 4-butylbenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6HQX
 
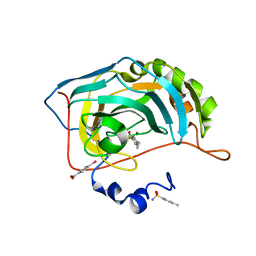 | | Human Carbonic Anhydrase II in complex with 4-Ethylbenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-ethylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6EH0
 
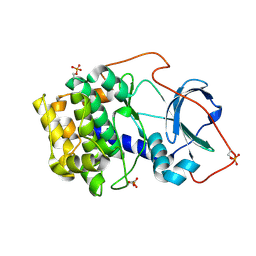 | |
6I2F
 
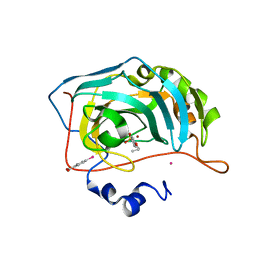 | | Human Carbonic Anhydrase II in complex with 4-Propoxybenzenesulfonamide | | Descriptor: | 4-propoxybenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6I1U
 
 | | Human Carbonic Anhydrase II in complex with 4-Ethoxybenzenesulfonamide | | Descriptor: | 4-ethoxybenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6I3E
 
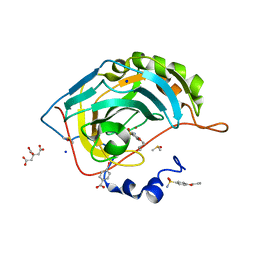 | | Human Carbonic Anhydrase II in complex with 4-Butylbenzenesulfonamide | | Descriptor: | 4-butoxybenzenesulfonamide, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6ERW
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp013 and Fasudil | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, UPF0418 protein FAM164A, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-10-19 | | Release date: | 2018-10-31 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
4Y4B
 
 | | Endothiapepsin in complex with fragment 323 | | Descriptor: | 6,6,8-trimethyl-5,6-dihydro[1,3]dioxolo[4,5-g]quinoline, ACETATE ION, Endothiapepsin, ... | | Authors: | Fu, K, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.108 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6EH3
 
 | |
6EH2
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp032 and AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
6EGW
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp017 and RKp117 | | Descriptor: | [2-[(4-isoquinolin-5-ylsulfonyl-1,4-diazepan-1-yl)methyl]phenyl]boronic acid, alpha-D-ribofuranose, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
6ERU
 
 | |
6I51
 
 | | Thrombin in complex with fragment J02 | | Descriptor: | 1H-isoindol-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Abazi, N, Heine, A, Klebe, G. | | Deposit date: | 2018-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment screening of Thrombin using a 96 member fragment library
To Be Published
|
|
6EM2
 
 | | Crystal structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with Fasudil | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-10-01 | | Release date: | 2018-10-10 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
6EMA
 
 | |
6ERV
 
 | |
6ERT
 
 | |
4E7R
 
 | | Thrombin in complex with 3-amidinophenylalanine inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-{[(2',4'-dichlorobiphenyl-3-yl)sulfonyl]amino}-3-oxopropyl]benzenecarboximidamide, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2012-03-19 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New 3-amidinophenylalanine-derived inhibitors of matriptase
MEDCHEMCOMM, 3, 2012
|
|
5HS6
 
 | | Human 17beta-hydroxysteroid dehydrogenase type 14 in complex with Estrone | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-01-25 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | New Insights into Human 17 beta-Hydroxysteroid Dehydrogenase Type 14: First Crystal Structures in Complex with a Steroidal Ligand and with a Potent Nonsteroidal Inhibitor.
J.Med.Chem., 59, 2016
|
|
6F14
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with isoquinoline and PKI (5-24) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOQUINOLINE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2017-11-21 | | Release date: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
5ICM
 
 | | 17beta-hydroxysteroid dehydrogenase type 14 in complex with a non-steroidal inhibitor | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | New Insights into Human 17 beta-Hydroxysteroid Dehydrogenase Type 14: First Crystal Structures in Complex with a Steroidal Ligand and with a Potent Nonsteroidal Inhibitor.
J.Med.Chem., 59, 2016
|
|
4N5P
 
 | | Thermolysin in complex with UBTLN20 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2013-10-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Methyl, Ethyl, Propyl, Butyl: Futile But Not for Water, as the Correlation of Structure and Thermodynamic Signature Shows in a Congeneric Series of Thermolysin Inhibitors.
Chemmedchem, 4, 2014
|
|
