3KS4
 
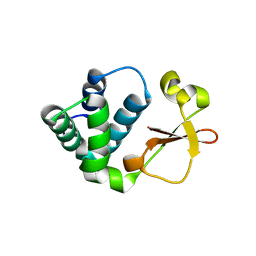 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
6UUP
 
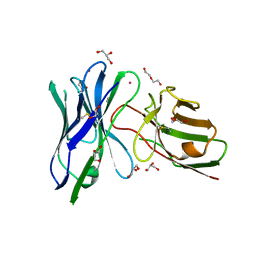 | | Structure of anti-hCD33 conditional scFv | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-CD33 conditional scFv, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kimberlin, C.R, Park, S. | | Deposit date: | 2019-10-31 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.20001864 Å) | | Cite: | Direct control of CAR T cells through small molecule-regulated antibodies.
Nat Commun, 12, 2021
|
|
6UY3
 
 | |
3KS8
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
6XKR
 
 | | Structure of Sasanlimab Fab in complex with PD-1 | | Descriptor: | GLYCEROL, Programmed cell death protein 1, Sasanlimab Fab Heavy chain, ... | | Authors: | Kimberlin, C.R, Chin, S.M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Pharmacologic Properties and Preclinical Activity of Sasanlimab, A High-affinity Engineered Anti-Human PD-1 Antibody.
Mol.Cancer Ther., 19, 2020
|
|
6MGE
 
 | | Structure of human 4-1BBL | | Descriptor: | GLYCEROL, PHOSPHATE ION, Tumor necrosis factor ligand superfamily member 9 | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6MHR
 
 | | Structure of the human 4-1BB / Urelumab Fab complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6MI2
 
 | | Structure of the human 4-1BB / Utomilumab Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6MGP
 
 | | Structure of human 4-1BB / 4-1BBL complex | | Descriptor: | ACETATE ION, GLYCEROL, Tumor necrosis factor ligand superfamily member 9, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab
Nat Commun, 9, 2018
|
|
6O2W
 
 | |
6O2U
 
 | | Crystal structure of the SARAF luminal domain | | Descriptor: | Store-operated calcium entry-associated regulatory factor | | Authors: | Kimberlin, C.R, Minor, D.L. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARAF Luminal Domain Structure Reveals a Novel Domain-Swapped beta-Sandwich Fold Important for SOCE Modulation.
J.Mol.Biol., 431, 2019
|
|
6O2V
 
 | |
3Q7C
 
 | | Exonuclease domain of Lassa virus nucleoprotein bound to manganese | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, PHOSPHATE ION, ... | | Authors: | Hastie, K.M, Kimberlin, C.R, Zandonatti, M.A, MacRae, I.J, Saphire, E.O. | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q7B
 
 | | Exonuclease domain of Lassa virus nucleoprotein | | Descriptor: | Nucleoprotein, PHOSPHATE ION, ZINC ION | | Authors: | Hastie, K.M, Kimberlin, C.R, Zandonatti, M.A, MacRae, I.J, Saphire, E.O. | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
