8ISR
 
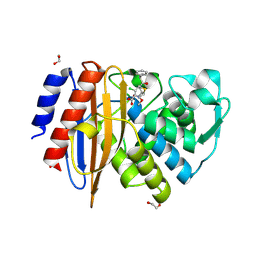 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
6SDF
 
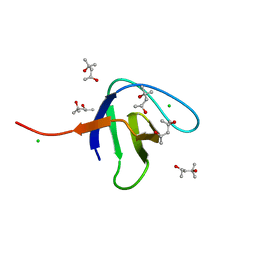 | | N-terminal SH3 domain of Grb2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Growth factor receptor-bound protein 2 | | Authors: | Bolgov, A.A, Korban, S.A, Luzik, D.A, Rogacheva, O.N, Zhemkov, V.A, Kim, M, Skrynnikov, N.R, Bezprozvanny, I.B. | | Deposit date: | 2019-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the SH3 domain of growth factor receptor-bound protein 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2UZ8
 
 | | The crystal structure of p18, human translation elongation factor 1 epsilon 1 | | Descriptor: | EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1, GLYCEROL | | Authors: | Kang, B.S, Kim, K.J, Kim, M.H, Oh, Y.S, Kim, S. | | Deposit date: | 2007-04-26 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of Three-Dimensional Structure and Residues of the Novel Tumor Suppressor Aimp3/P18 Required for the Interaction with Atm.
J.Biol.Chem., 283, 2008
|
|
2EWT
 
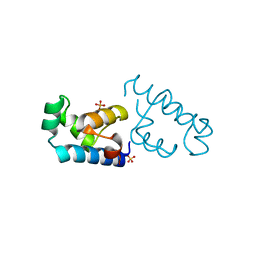 | | Crystal structure of the DNA-binding domain of BldD | | Descriptor: | SULFATE ION, putative DNA-binding protein | | Authors: | Kim, I.K, Lee, C.J, Kim, M.K, Kim, J.M, Kim, J.H, Yim, H.S, Cha, S.S, Kang, S.O. | | Deposit date: | 2005-11-07 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2)
Mol.Microbiol., 60, 2006
|
|
2ZCU
 
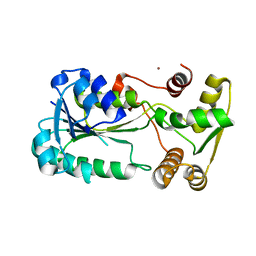 | | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from escherichia coli | | Descriptor: | COPPER (II) ION, Uncharacterized oxidoreductase ytfG | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
2XMQ
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
2XMR
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
2XMS
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | CHLORIDE ION, IMIDAZOLE, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
2ZCV
 
 | | Crystal structure of NADPH-dependent quinone oxidoreductase QOR2 complexed with NADPH from escherichia coli | | Descriptor: | COPPER (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
7XP3
 
 | | DNA complex form of ORESARA1(ANAC092) NAC Domain | | Descriptor: | DNA (5'-D(*AP*GP*TP*TP*AP*CP*GP*TP*AP*CP*GP*GP*CP*AP*CP*AP*CP*GP*TP*AP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*CP*GP*TP*GP*TP*GP*CP*CP*GP*TP*AP*CP*GP*TP*AP*AP*C)-3'), DNA (5'-D(P*CP*AP*CP*AP*CP*GP*TP*AP*AP*C)-3'), ... | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2022-05-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of DNA binding by the NAC transcription factor ORE1, a master regulator of plant senescence.
Plant Commun., 4, 2023
|
|
2B8I
 
 | | Crystal Structure and Functional Studies Reveal that PAS Factor from Vibrio vulnificus is a Novel Member of the Saposin-Fold Family | | Descriptor: | PAS factor | | Authors: | Lee, J.H, Yang, S.T, Rho, S.H, Im, Y.J, Kim, S.Y, Kim, Y.R, Kim, M.K, Kang, G.B, Kim, J.I, Rhee, J.H, Eom, S.H. | | Deposit date: | 2005-10-07 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and functional studies reveal that PAS factor from Vibrio vulnificus is a novel member of the saposin-fold family
J.Mol.Biol., 355, 2006
|
|
5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | Descriptor: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
2RFG
 
 | |
2VRB
 
 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIPHENYLMETHANE REDUCTASE | | Authors: | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | Deposit date: | 2008-03-31 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
2VRC
 
 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | Descriptor: | TRIPHENYLMETHANE REDUCTASE | | Authors: | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | Deposit date: | 2008-03-31 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
2Z99
 
 | | Crystal Structure of ScpB from Mycobacterium tuberculosis | | Descriptor: | Putative uncharacterized protein | | Authors: | Kim, J.-S, Lee, S, Kang, B.S, Kim, M.H, Lee, H.-S, Kim, K.J. | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and domain characterization of ScpB from Mycobacterium tuberculosis
Proteins, 71, 2008
|
|
2CB0
 
 | |
3CRA
 
 | | Crystal Structure of Escherichia coli MazG, the Regulator of Nutritional Stress Response | | Descriptor: | Protein mazG | | Authors: | Lee, S, Kim, M.H, Kang, B.S, Kim, J.S, Kim, Y.G, Kim, K.J. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
J.Biol.Chem., 283, 2008
|
|
3CRC
 
 | | Crystal Structure of Escherichia coli MazG, the Regulator of Nutritional Stress Response | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein mazG | | Authors: | Lee, S, Kim, M.H, Kang, B.S, Kim, J.S, Kim, Y.G, Kim, K.J. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
J.Biol.Chem., 283, 2008
|
|
8SLU
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and high pressure (205 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SLS
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and ambient pressure (0.1 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SLT
 
 | | Crystal structure of human STEP (PTPN5) at physiological temperature (310 K) and ambient pressure (0.1 MPa) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
1FYJ
 
 | | SOLUTION STRUCTURE OF MULTI-FUNCTIONAL PEPTIDE MOTIF-1 PRESENT IN HUMAN GLUTAMYL-PROLYL TRNA SYNTHETASE (EPRS). | | Descriptor: | MULTIFUNCTIONAL AMINOACYL-TRNA SYNTHETASE | | Authors: | Jeong, E.-J, Hwang, G.-S, Kim, K.H, Kim, M.J, Kim, S, Kim, K.-S. | | Deposit date: | 2000-10-02 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of multifunctional peptide motifs in human bifunctional tRNA synthetase: identification of RNA-binding residues and functional implications for tandem repeats.
Biochemistry, 39, 2000
|
|
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
