8I07
 
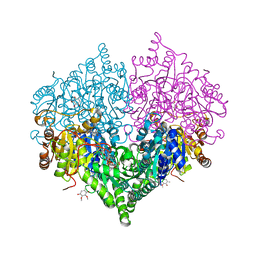 | |
8I08
 
 | | Crystal structure of Escherichia coli glyoxylate carboligase quadruple mutant | | 分子名称: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | 著者 | Kim, J.H, Kim, J.S. | | 登録日 | 2023-01-10 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
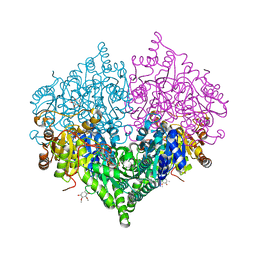
8I01
 
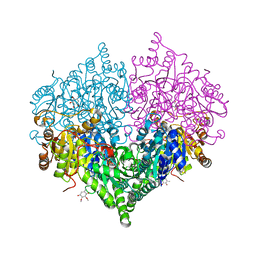 | | Crystal structure of Escherichia coli glyoxylate carboligase | | 分子名称: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | 著者 | Kim, J.H, Kim, J.S. | | 登録日 | 2023-01-10 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8I05
 
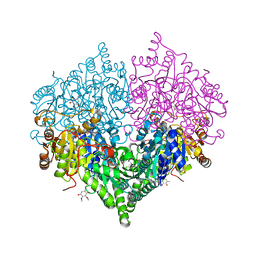 | | Crystal structure of Escherichia coli glyoxylate carboligase double mutant | | 分子名称: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | 著者 | Kim, J.H, Kim, J.S. | | 登録日 | 2023-01-10 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
5X1E
 
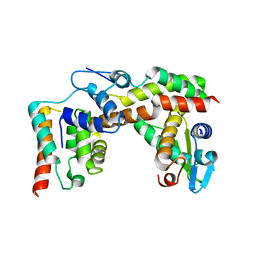 | |
5HZ2
 
 | | Crystal structure of PhaC1 from Ralstonia eutropha | | 分子名称: | GLYCEROL, Poly-beta-hydroxybutyrate polymerase, SULFATE ION | | 著者 | Kim, J, Kim, K.-J. | | 登録日 | 2016-02-02 | | 公開日 | 2016-12-07 | | 最終更新日 | 2017-04-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms.
Biotechnol J, 12, 2017
|
|
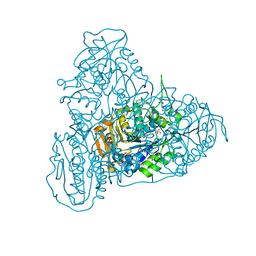
7YQ3
 
 | | human insulin receptor bound with A43 DNA aptamer and insulin | | 分子名称: | IR-A43 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
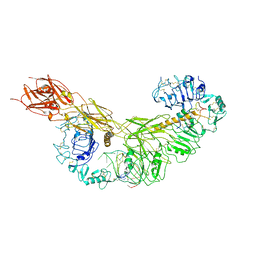
7YQ4
 
 | | human insulin receptor bound with A62 DNA aptamer and insulin - locally refined | | 分子名称: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
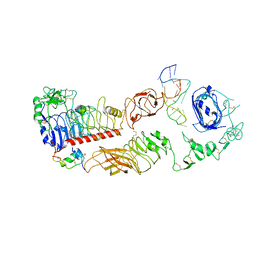
7YQ6
 
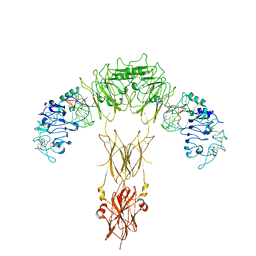 | | human insulin receptor bound with A62 DNA aptamer | | 分子名称: | IR-A62 aptamer, Isoform Short of Insulin receptor | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ5
 
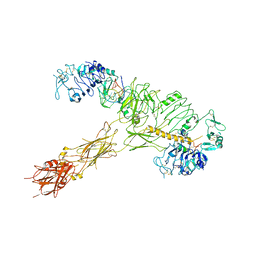 | | human insulin receptor bound with A62 DNA aptamer and insulin | | 分子名称: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.27 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
5WE1
 
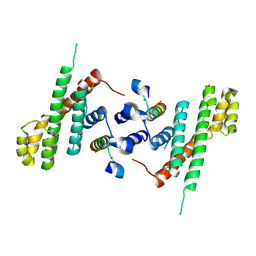 | | Structural Basis for Shelterin Bridge Assembly | | 分子名称: | Protection of telomeres protein poz1,Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ZINC ION | | 著者 | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | 登録日 | 2017-07-06 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.202 Å) | | 主引用文献 | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
3MDD
 
 | |
4GEK
 
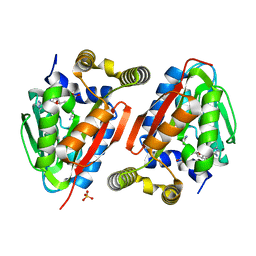 | | Crystal Structure of wild-type CmoA from E.coli | | 分子名称: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA (cmo5U34)-methyltransferase | | 著者 | Kim, J, Toro, R, Bonanno, J.B, Bhosle, R, Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-08-02 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Nature, 498, 2013
|
|
4PTS
 
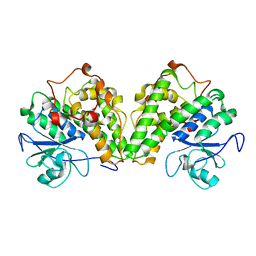 | | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405 | | 分子名称: | glutathione S-transferase | | 著者 | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-03-11 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405
To be Published
|
|
4PUA
 
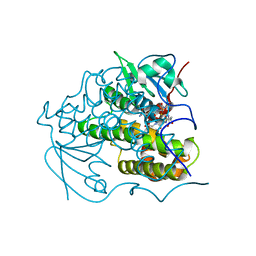 | | Crystal Structure Of glutathione transferase YghU from Streptococcus pneumoniae ATCC 700669, complexed with glutathione, Target EFI-507284 | | 分子名称: | GLUTATHIONE, glutathione S-transferase | | 著者 | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-03-12 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.708 Å) | | 主引用文献 | Crystal Structure Of glutathione transferase YghU from Streptococcus pneumoniae ATCC 700669, complexed with glutathione, Target EFI-507284
To be Published
|
|
7YMG
 
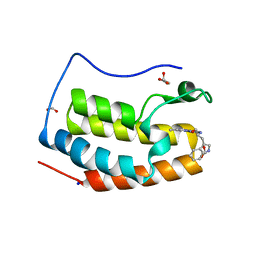 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 2-({3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl}amino)-3-(1H-indol-3-yl)propan-1-ol | | 分子名称: | (2S)-2-[(3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)amino]-3-(1H-indol-3-yl)propan-1-ol, Bromodomain-containing protein 4, FORMIC ACID, ... | | 著者 | Kim, J.H, Lee, B.I. | | 登録日 | 2022-07-28 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
7YQ9
 
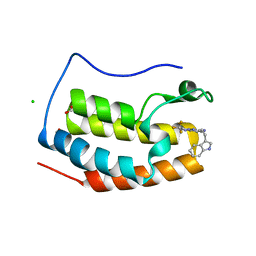 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N-[2-(1H-indol-3-yl)ethyl]-3-(trifluoromethyl)[1,2,4]triazolo[4,3-b]pyridazin-6-amine | | 分子名称: | Bromodomain-containing protein 4, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Kim, J.H, Lee, B.I. | | 登録日 | 2022-08-05 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
7CXT
 
 | |
6L5H
 
 | |
6L5J
 
 | |
6U7V
 
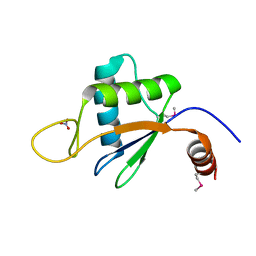 | | xRRM structure of spPof8 | | 分子名称: | NITRATE ION, Protein pof8 | | 著者 | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | 登録日 | 2019-09-03 | | 公開日 | 2020-09-09 | | 最終更新日 | 2021-03-24 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
7CT6
 
 | |
6UKJ
 
 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | 著者 | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | 登録日 | 2019-10-05 | | 公開日 | 2019-12-04 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
8K6X
 
 | | Crystal structure of E.coli Cyanase complex with cyanate and bicarbonate | | 分子名称: | CARBONATE ION, Cyanate hydratase, SULFATE ION, ... | | 著者 | Kim, J, Nam, K.H, Cho, Y. | | 登録日 | 2023-07-25 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural mechanism of Escherichia coli cyanase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8K6U
 
 | |
