7DGQ
 
 | | Activity optimized supercomplex state1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DKF
 
 | | Activity optimized supercomplex state4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
6OIC
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q (sulfite soaked) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, Sulfide:quinone oxidoreductase, ... | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
4HY2
 
 | |
5ZXB
 
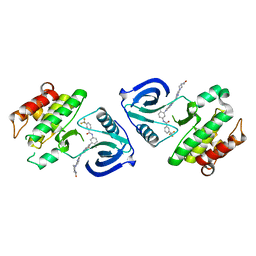 | | Crystal structure of ACK1 with compound 10d | | Descriptor: | Activated CDC42 kinase 1, N-{3-[7-{[6-(4-acetylpiperazin-1-yl)pyridin-3-yl]amino}-1-methyl-2-oxo-1,4-dihydropyrimido[4,5-d]pyrimidin-3(2H)-yl]-4-methylphenyl}-3-(trifluoromethyl)benzamide | | Authors: | Hong, E.M, Kim, H.L, Sim, T.B. | | Deposit date: | 2018-05-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | First SAR Study for Overriding NRAS Mutant Driven Acute Myeloid Leukemia.
J. Med. Chem., 61, 2018
|
|
1K1D
 
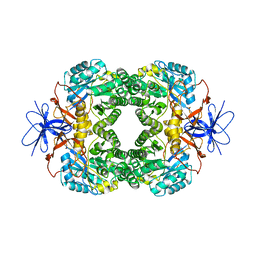 | | Crystal structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Cheon, Y.H, Kim, H.S, Han, K.H, Abendroth, J, Niefind, K, Schomburg, D, Wang, J, Kim, Y. | | Deposit date: | 2001-09-25 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of D-hydantoinase from Bacillus stearothermophilus: insight into the stereochemistry of enantioselectivity.
Biochemistry, 41, 2002
|
|
8I9Q
 
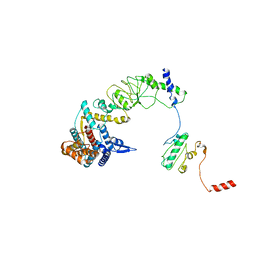 | | The focused refinement of CCT4-PhLP2A from TRiC-PhLP2A complex in the open state | | Descriptor: | Phosducin-like protein 3, T-complex protein 1 subunit delta | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I6J
 
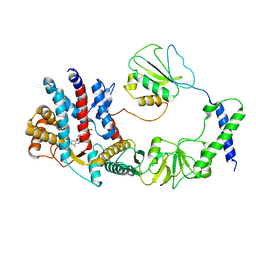 | | The focused refinement of CCT3-PhLP2A from TRiC-PhLP2A complex in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosducin-like protein 3, T-complex protein 1 subunit gamma | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
3QD6
 
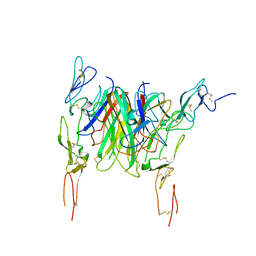 | | Crystal structure of the CD40 and CD154 (CD40L) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Lee, J.-O, Kim, Y.J, Song, D.H, Kim, H.M, Park, B.S. | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation
J.Biol.Chem., 286, 2011
|
|
4RCP
 
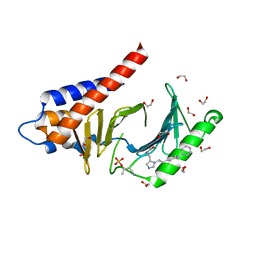 | | Crystal structure of Plk1 Polo-box domain in complex with PL-2 | | Descriptor: | 1,2-ETHANEDIOL, PL-2, Serine/threonine-protein kinase PLK1 | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-12 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
4OH9
 
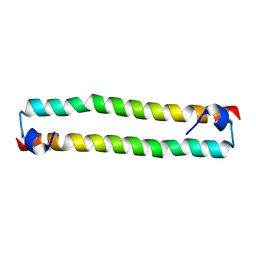 | | Crystal Structure of the human MST2 SARAH homodimer | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OH8
 
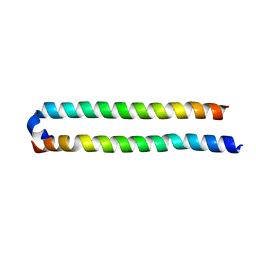 | | Crystal Structure of the human MST1-RASSF5 SARAH heterodimer | | Descriptor: | Ras association domain-containing protein 5, Serine/threonine-protein kinase 4 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4GCV
 
 | |
1Y4S
 
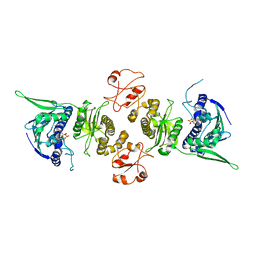 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
1Y4U
 
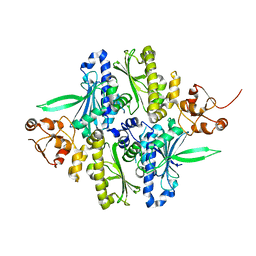 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
1KU0
 
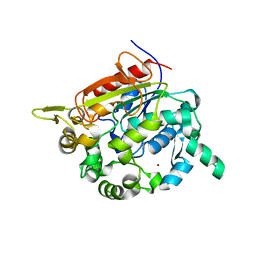 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
7ML1
 
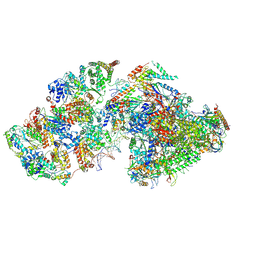 | | RNA polymerase II pre-initiation complex (PIC2) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML2
 
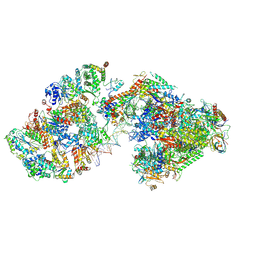 | | RNA polymerase II pre-initiation complex (PIC3) | | Descriptor: | BJ4_G0004860.mRNA.1.CDS.1, BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML4
 
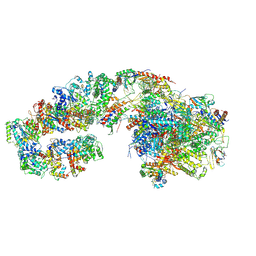 | | RNA polymerase II initially transcribing complex (ITC) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML0
 
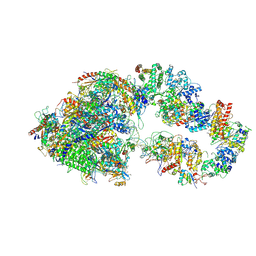 | | RNA polymerase II pre-initiation complex (PIC1) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7KPW
 
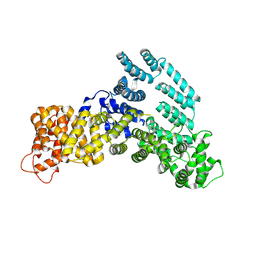 | | Structure of the H-lobe of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12 | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
4NUH
 
 | | Crystal structure of mLeIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ZG0
 
 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
7N99
 
 | | SDE2 SAP domain apo structure | | Descriptor: | Isoform 2 of Replication stress response regulator SDE2 | | Authors: | Paung, Y, Weinheimer, A.S, Rageul, J, Khan, A, Ho, B, Tong, M, Alphonse, S, Seeliger, M.A, Kim, H. | | Deposit date: | 2021-06-17 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Extended DNA-binding interfaces beyond the canonical SAP domain contribute to the function of replication stress regulator SDE2 at DNA replication forks.
J.Biol.Chem., 298, 2022
|
|
4NU3
 
 | | Crystal structure of mFfIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | SODIUM ION, SULFATE ION, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
