7VCJ
 
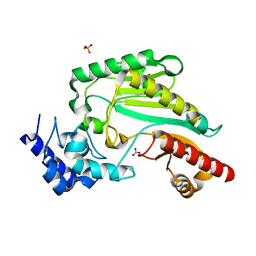 | | Arginine kinase H227A from Daphnia magna | | Descriptor: | Arginine kinase, NITRATE ION, PHOSPHATE ION | | Authors: | Kim, D.S, Jang, K, Kim, W.S, Kim, Y.J, Park, J.H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability.
Molecules, 27, 2022
|
|
9ISA
 
 | |
7C04
 
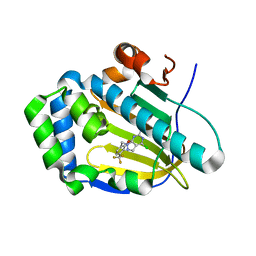 | | Crystal structure of human Trap1 with DN203492 | | Descriptor: | 4-chloranyl-1-[[2-methoxy-4-(trifluoromethyl)phenyl]methyl]pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
7C05
 
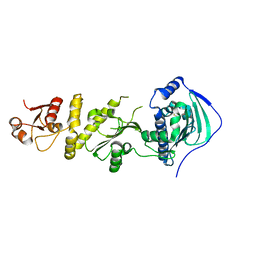 | | Crystal structure of human Trap1 with DN203495 | | Descriptor: | 1-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-4-chloranyl-pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
4HVZ
 
 | |
7UPH
 
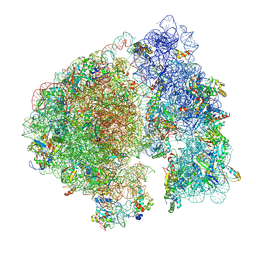 | | Structure of a ribosome with tethered subunits | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kim, D.S, Watkins, A, Bidstrup, E, Lee, J, Topkar, V.V, Kofman, C, Schwarz, K.J, Liu, Y, Pintilie, G, Roney, E, Das, R, Jewett, M.C. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-17 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Three-dimensional structure-guided evolution of a ribosome with tethered subunits.
Nat.Chem.Biol., 18, 2022
|
|
7C7B
 
 | | Crystal structure of human TRAP1 with SJT009 | | Descriptor: | 2-azanyl-9-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
7C7C
 
 | | Crystal structure of human TRAP1 with SJT104 | | Descriptor: | 2-azanyl-9-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
4KMF
 
 | |
6JKG
 
 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
6JKH
 
 | | The NAD+-bound form of human NSDHL | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
8X3P
 
 | |
8X3O
 
 | |
3IQY
 
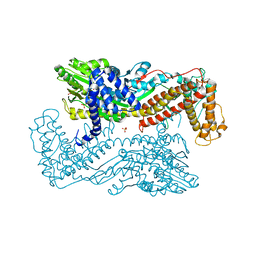 | | Active site mutants of B. subtilis SecA | | Descriptor: | Protein translocase subunit secA, SULFATE ION | | Authors: | Kim, D, Hunt, J.F. | | Deposit date: | 2009-08-21 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ATPase Active-Site Electrostatic Interactions Control the Global Conformation of the 100 kDa SecA Translocase.
J.Am.Chem.Soc., 135, 2013
|
|
3IQM
 
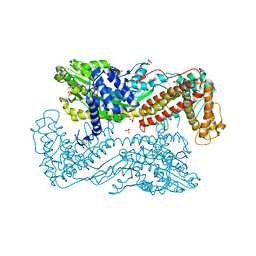 | | Active site mutants of B. subtilis SecA | | Descriptor: | Protein translocase subunit secA, SULFATE ION | | Authors: | Kim, D, Hunt, J.F. | | Deposit date: | 2009-08-20 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ATPase Active-Site Electrostatic Interactions Control the Global Conformation of the 100 kDa SecA Translocase.
J.Am.Chem.Soc., 135, 2013
|
|
1X70
 
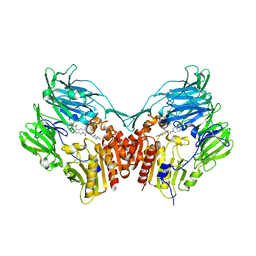 | | HUMAN DIPEPTIDYL PEPTIDASE IV IN COMPLEX WITH A BETA AMINO ACID INHIBITOR | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, D, Wang, L, Beconi, M, Eiermann, G.J, Fisher, M.H, He, H, Hickey, G.J, Leiting, B, Lyons, K. | | Deposit date: | 2004-08-12 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (2R)-4-Oxo-4-[3-(Trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin- 7(8H)-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: A Potent, Orally Active Dipeptidyl Peptidase IV Inhibitor for the Treatment of Type 2 Diabetes
J.Med.Chem., 48, 2005
|
|
3M9Q
 
 | |
3OA6
 
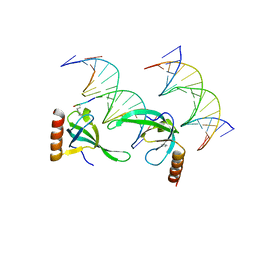 | | Human MSL3 Chromodomain bound to DNA and H4K20me1 peptide | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), H4 peptide monomethylated at lysine 20, ... | | Authors: | Kim, D, Huang, P, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3P7J
 
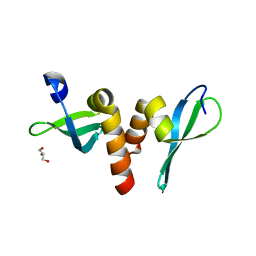 | | Drosophila HP1a chromo shadow domain | | Descriptor: | GLYCEROL, Heterochromatin protein 1 | | Authors: | Kim, D, Chruszcz, M, Minor, W, Khorasanizadeh, S. | | Deposit date: | 2010-10-12 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The HP1a Disordered C Terminus and Chromo Shadow Domain Cooperate to Select Target Peptide Partners.
Chembiochem, 12, 2011
|
|
2N8N
 
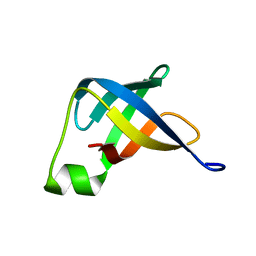 | | Solution structure of translation initiation factor | | Descriptor: | Translation initiation factor IF-1 | | Authors: | Kim, D, Lee, K, Lee, B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics study of translation initiation factor 1 from Staphylococcus aureus suggests its RNA binding mode
Biochim.Biophys.Acta, 1865, 2017
|
|
4OYH
 
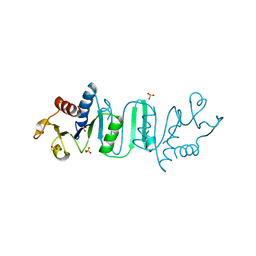 | | Structure of Bacillus subtilis MobB | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Kim, D, Choe, J. | | Deposit date: | 2014-02-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Bacillus subtilis MobB
To Be Published
|
|
5GUY
 
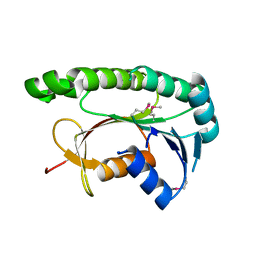 | | Structural and functional study of chuY from E. coli CFT073 strain | | Descriptor: | Uncharacterized protein chuY | | Authors: | Kim, H, Choi, J, HA, S.C, Chaurasia, A.K, Kim, D, Kim, K.K. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural and functional study of ChuY from Escherichia coli strain CFT073
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5XEQ
 
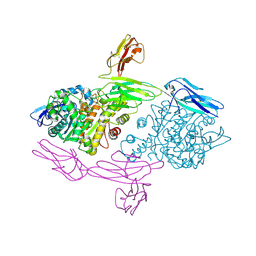 | | Crystal Structure of human MDGA1 and human neuroligin-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, ... | | Authors: | Kim, H.M, Kim, J.A, Kim, D. | | Deposit date: | 2017-04-05 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.136 Å) | | Cite: | Structural Insights into Modulation of Neurexin-Neuroligin Trans-synaptic Adhesion by MDGA1/Neuroligin-2 Complex
Neuron, 94, 2017
|
|
4NKR
 
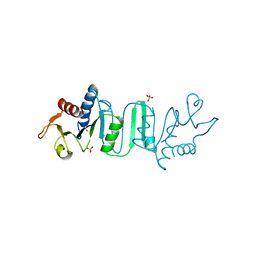 | | The Crystal structure of Bacillus subtilis MobB | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Choe, J, Kim, D, Choi, S, Kim, H. | | Deposit date: | 2013-11-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Insight into the Role of E.coli MobB in Molybdenum Cofactor Biosynthesis based on the high resolution crystal structure
TO BE PUBLISHED
|
|
5WY8
 
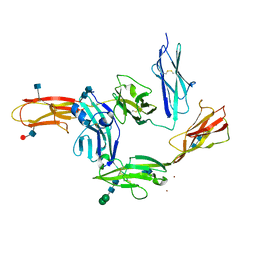 | | Crystal structure of PTP delta Ig1-Ig3 in complex with IL1RAPL1 Ig1-Ig3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Kim, H.M, Won, S.Y, Kim, D. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | LAR-RPTP Clustering Is Modulated by Competitive Binding between Synaptic Adhesion Partners and Heparan Sulfate
Front Mol Neurosci, 10, 2017
|
|
