6U8N
 
 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Fully extended filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U1S
 
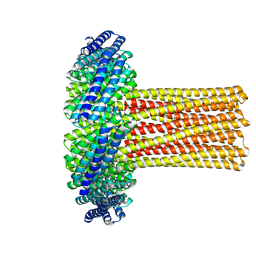 | | Cryo-EM structure of a de novo designed 16-helix transmembrane nanopore, TMHC8_R. | | Descriptor: | de novo designed 16-helix transmembrane nanopore, TMHC8_R | | Authors: | Johnson, M.J, Reggiano, G, Xu, C, Lu, P, Hsia, Y, Brunette, T.J, DiMaio, F, Baker, D, Kollman, J. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Computational Design of Transmembrane Channels
To Be Published
|
|
6UDQ
 
 | | Human IMPDH2 treated with ATP, IMP, and 20 mM GTP. Fully compressed filament end reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-19 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UDO
 
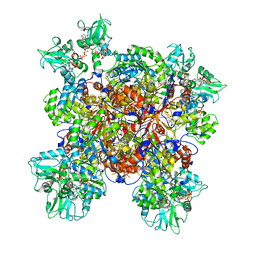 | | Human IMPDH2 treated with ATP, IMP, and 20 mM GTP. Fully compressed filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-19 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8E
 
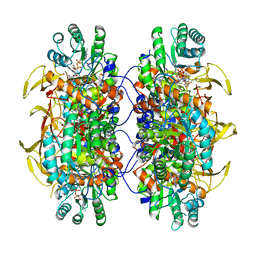 | |
6UDP
 
 | |
6U8R
 
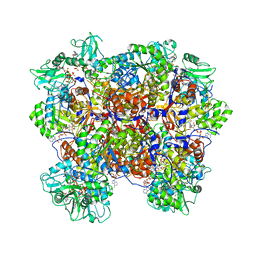 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Bent (1/4 compressed, 3/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UA4
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Bent (3/4 compressed, 1/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U9O
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Fully compressed filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
2AFE
 
 | | Solution Structure of Asl1650, an Acyl Carrier Protein from Anabaena sp. PCC 7120 with a Variant Phosphopantetheinylation-Site Sequence | | Descriptor: | protein Asl1650 | | Authors: | Johnson, M.A, Peti, W, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Asl1650, an acyl carrier protein from Anabaena sp. PCC 7120 with a variant phosphopantetheinylation-site sequence
Protein Sci., 15, 2006
|
|
2AFD
 
 | | Solution Structure of Asl1650, an Acyl Carrier Protein from Anabaena sp. PCC 7120 with a Variant Phosphopantetheinylation-Site Sequence | | Descriptor: | protein Asl1650 | | Authors: | Johnson, M.A, Peti, W, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Asl1650, an acyl carrier protein from Anabaena sp. PCC 7120 with a variant phosphopantetheinylation-site sequence
Protein Sci., 15, 2006
|
|
6UA5
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Free interfacial octamer reconstruction. | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8S
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Filament assembly interface reconstruction. | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UAJ
 
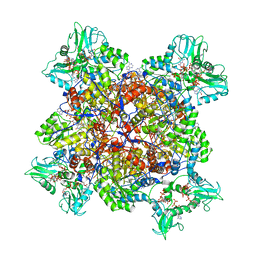 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Free canonical octamer reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UC2
 
 | |
6UA2
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Bent (2/4 compressed, 2/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
7UJD
 
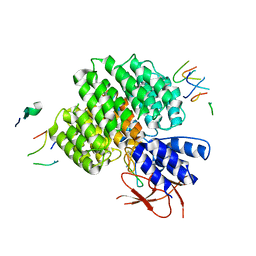 | | PSMD2 Structure bound to MC1 and Fab8/14 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, ACY-PHE-PRO-ASP-VAL-SAR-LEU-HIS-ARG-TYR-TRP-GLY-TRP-ASP-CYS-GLY-NH2, Fab 14 HC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
7UIH
 
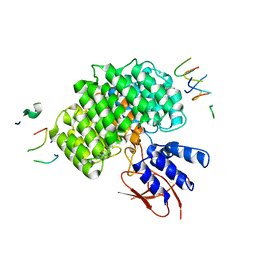 | | PSMD2 Structure | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, Fab 14 HC CDRs, Fab 14 LC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
2JNQ
 
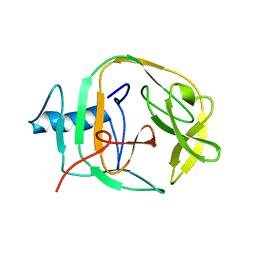 | | Solution Structure of a KlbA Intein Precursor from Methanococcus jannaschii | | Descriptor: | Hypothetical protein MJ0781 | | Authors: | Johnson, M.A, Southworth, M.W, Herrmann, T, Brace, L, Perler, F.B, Wuthrich, K.A. | | Deposit date: | 2007-01-31 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a KlbA intein precursor from Methanococcus jannaschii
Protein Sci., 16, 2007
|
|
2JMZ
 
 | | Solution structure of a KlbA intein precursor from Methanococcus jannaschii | | Descriptor: | Hypothetical protein MJ0781 | | Authors: | Johnson, M.A, Southworth, M.W, Herrmann, T, Brace, L, Perler, F.B, Wuthrich, K.A. | | Deposit date: | 2006-12-14 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a KlbA intein precursor from Methanococcus jannaschii
Protein Sci., 16, 2007
|
|
2KQV
 
 | |
2KAF
 
 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | Descriptor: | Non-structural protein 3 | | Authors: | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
2KQW
 
 | |
2KYS
 
 | | NMR Structure of the SARS Coronavirus Nonstructural Protein Nsp7 in Solution at pH 6.5 | | Descriptor: | Non-structural protein 7 | | Authors: | Johnson, M.A, Jaudzems, K, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the SARS-CoV Nonstructural Protein 7 in Solution at pH 6.5.
J.Mol.Biol., 402, 2010
|
|
6MEB
 
 | | Crystal structure of Tylonycteris bat coronavirus HKU4 macrodomain in complex with nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
