7L0A
 
 | |
8D38
 
 | | Structure of a purine nucleoside phosphorylase from Geobacillus stearothermophilus | | Descriptor: | Purine nucleoside phosphorylase, SODIUM ION | | Authors: | Given, F, Johnston, J, Crittenden, D, Moran, F, Johns, A. | | Deposit date: | 2022-05-31 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The structure of His-tagged Geobacillus stearothermophilus purine nucleoside phosphorylase reveals a 'spanner in the works'.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8SM3
 
 | | Structure of Bacillus cereus VD045 Gabija GajA-GajB Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB, SULFATE ION | | Authors: | Antine, S.P, Mooney, S.E, Johnson, A.G, Kranzusch, P.J. | | Deposit date: | 2023-04-25 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
1FMG
 
 | | CRYSTAL STRUCTURE OF PORCINE BETA TRYPSIN WITH 0.04% POLYDOCANOL | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Deepthi, S, Johnson, A, Pattabhi, V. | | Deposit date: | 2000-08-17 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of porcine beta-trypsin-detergent complexes: the stabilization of proteins through hydrophilic binding of polydocanol.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FNI
 
 | | CRYSTAL STRUCTURE OF PORCINE BETA TRYPSIN WITH 0.01% POLYDOCANOL | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Deepthi, S, Johnson, A, Pattabhi, V. | | Deposit date: | 2000-08-22 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of porcine beta-trypsin-detergent complexes: the stabilization of proteins through hydrophilic binding of polydocanol.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FN6
 
 | | CRYSTAL STRUCTURE OF PORCINE BETA TRYPSIN WITH 0.1% POLYDOCANOL | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, METHANOL, ... | | Authors: | Deepthi, S, Johnson, A, Pattabhi, V. | | Deposit date: | 2000-08-21 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of porcine beta-trypsin-detergent complexes: the stabilization of proteins through hydrophilic binding of polydocanol.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3G0X
 
 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 5 | | Descriptor: | (2Z)-N-biphenyl-4-yl-2-cyano-3-cyclopropyl-3-hydroxyprop-2-enamide, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Davies, M, Heikkila, T, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
2RLU
 
 | | The Three Dimensional Structure of the Moorella thermoacetica Selenocysteine Insertion Sequence RNA Hairpin and its Interaction with the Elongation factor SelB | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*GP*CP*GP*GP*GP*UP*CP*UP*CP*GP*CP*AP*AP*CP*C)-3') | | Authors: | Beribisky, A.V, Tavares, T.J, Amborski, A.N, Motamed, M, Johnson, A.E, Mark, T.L, Johnson, P.E. | | Deposit date: | 2007-08-21 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the Moorella thermoacetica selenocysteine insertion sequence RNA hairpin and its interaction with the elongation factor SelB
Rna, 13, 2007
|
|
1APL
 
 | | CRYSTAL STRUCTURE OF A MAT-ALPHA2 HOMEODOMAIN-OPERATOR COMPLEX SUGGESTS A GENERAL MODEL FOR HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*C P*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT-ALPHA2 HOMEODOMAIN) | | Authors: | Wolberger, C, Vershon, A.K, Liu, B, Johnson, A.D, Pabo, C.O. | | Deposit date: | 1993-10-04 | | Release date: | 1993-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 67, 1991
|
|
4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
1ERJ
 
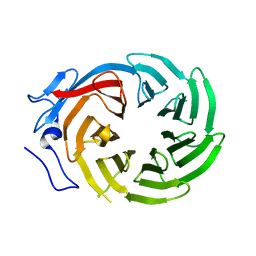 | | CRYSTAL STRUCTURE OF THE C-TERMINAL WD40 DOMAIN OF TUP1 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR TUP1 | | Authors: | Sprague, E.R, Redd, M.J, Johnson, A.D, Wolberger, C. | | Deposit date: | 2000-04-06 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast.
EMBO J., 19, 2000
|
|
1YRN
 
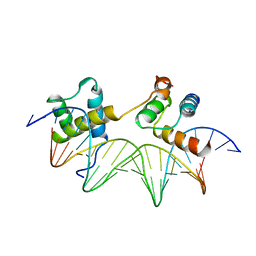 | | CRYSTAL STRUCTURE OF THE MATA1/MATALPHA2 HOMEODOMAIN HETERODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*TP*AP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*TP*AP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT A1 HOMEODOMAIN), ... | | Authors: | Li, T, Stark, M.R, Johnson, A.D, Wolberger, C. | | Deposit date: | 1995-11-02 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA.
Science, 270, 1995
|
|
5T62
 
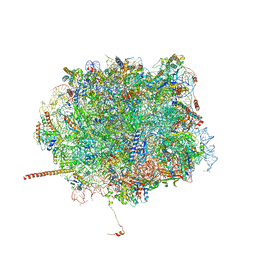 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3-Tif6-Lsg1 Complex | | Descriptor: | 5.8S Ribosomal RNA, 5S Ribosomal RNA, 60S ribosomal export protein NMD3, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
7VJY
 
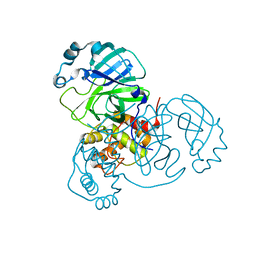 | |
8U7I
 
 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
8FNW
 
 | | Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, Archaeal ATPase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNT
 
 | | Structure of RdrA from Escherichia coli RADAR defense system | | Descriptor: | Archaeal ATPase | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNV
 
 | | Structure of RdrB from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | Descriptor: | KAP NTPase domain-containing protein | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
7UG6
 
 | | Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Yelland, J.N, Bravo, J.P.K, Black, J.J.B, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2022-03-24 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A single 2'-O-methylation of ribosomal RNA gates assembly of a functional ribosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6N8K
 
 | | Cryo-EM structure of early cytoplasmic-immediate (ECI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8O
 
 | | Cryo-EM structure of Rpl10-inserted (RI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8J
 
 | | Cryo-EM structure of late nuclear (LN) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal protein L11-A, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8L
 
 | | Cryo-EM structure of early cytoplasmic-late (ECL) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8M
 
 | | Cryo-EM structure of pre-Lsg1 (PL) pre-60S ribosomal subunit | | Descriptor: | 5.8S RNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
