6YQP
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW22 | | Descriptor: | (~{E})-3-[4-[[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6 ),4,7,10,12-pentaen-9-yl]ethanoylamino]methyl]phenyl]-~{N}-oxidanyl-prop-2-enamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
1DZX
 
 | |
1DZW
 
 | |
6YQO
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW12 | | Descriptor: | (S)-N1-(4-(2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetamido)phenyl)-N8-hydroxyoctanediamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
7BE4
 
 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR159 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7BE5
 
 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR276 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-methyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8000524 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
2XWR
 
 | |
2WQJ
 
 | |
4CZ5
 
 | |
4CZ7
 
 | |
4CZ6
 
 | |
8CG7
 
 | | Structure of p53 cancer mutant Y220C with arylation at Cys182 and Cys277 | | Descriptor: | 1,2-ETHANEDIOL, 5-iodanyl-2-methylsulfonyl-pyrimidine, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Pichon, M.M, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Reactivity Studies of 2-Sulfonylpyrimidines Allow Selective Protein Arylation.
Bioconjug.Chem., 34, 2023
|
|
8A31
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC694 | | Descriptor: | 4-(3-fluoranylpyrrol-1-yl)-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8A32
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC769 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3,4-bis(fluoranyl)pyrrol-1-yl]-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
6ZNV
 
 | | Protein polybromo-1 (PB1 BD2) Bound To DP28 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperidin-4-ol, ACETATE ION, ... | | Authors: | Preuss, F, Mathea, S, Chatterjee, D, Wanior, M, Joerger, A.C, Knapp, S. | | Deposit date: | 2020-07-06 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6ZS2
 
 | | Crystal Structure of the bromodomain of human transcription activator BRG1 (SMARCA4) in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Transcription activator BRG1 | | Authors: | Preuss, F, Joerger, A.C, Kraemer, A, Wanior, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6ZS4
 
 | | Crystal structure of the fifth bromodomain of human protein polybromo-1 in complex with tert-butyl 4-[3-amino-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazine-1-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, Protein polybromo-1, tert-butyl 4-[3-amino-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazine-1-carboxylate | | Authors: | Preuss, F, Joerger, A.C, Kraemer, A, Wanior, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
7Q8A
 
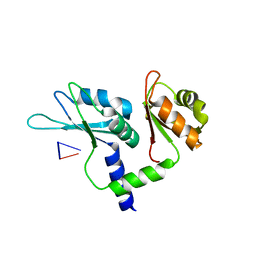 | | Crystal structure of tandem domain RRM1-2 of FUBP-interacting repressor (FIR) bound to FUSE ssDNA fragment | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*GP*T)-3'), Poly(U)-binding-splicing factor PUF60, ... | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tandem domain RRM1-2 of FIR bound to FUSE ssDNA fragment
To Be Published
|
|
9F1M
 
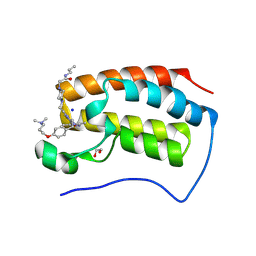 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 45 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-[7-[2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanoylamino]heptanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.90001261 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 45
to be published
|
|
9F1N
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 46 | | Descriptor: | (2S,4R)-1-[(2S)-2-[9-[2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanoylamino]nonanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-4H-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.71000016 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 46
to be published
|
|
9F1J
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 14 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[2-[2-[2-[2-[2-[2-[2,6-bis(oxidanylidene)piperidin-3-yl]-1,3-bis(oxidanylidene)isoindol-4-yl]oxyethanoylamino]ethoxy]ethoxy]ethoxy]ethyl]-4-[4-[2-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(2-morpholin-4-ylethyl)benzimidazol-2-yl]ethyl]phenoxy]butanamide | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.13003039 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 14
to be published
|
|
9F1L
 
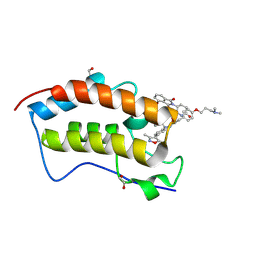 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 35 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[2-[2-[(3R)-2,6-bis(oxidanylidene)piperidin-3-yl]-1,3-bis(oxidanylidene)isoindol-4-yl]oxyethyl]-2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanamide | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.30000627 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 35
to be published
|
|
9F1K
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based inhibitor 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]-N-(2-methoxyethyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based inhibitor 30
to be published
|
|
7PC6
 
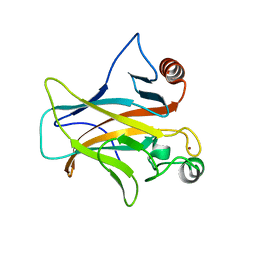 | | DNA-binding domain of a p53 homolog from the hydrothermal vent annelid Alvinella pompejana | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding domain, ZINC ION | | Authors: | Balourdas, D.-I, Knapp, S, Soussi, T, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolutionary history of the p53 family DNA-binding domain: insights from an Alvinella pompejana homolog.
Cell Death Dis, 13, 2022
|
|
8BI8
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase | | Descriptor: | Nitric oxide synthase, brain | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
