4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
5XG5
 
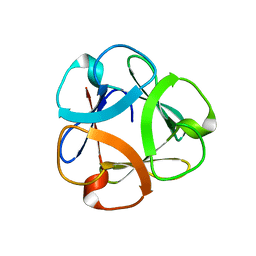 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
6QSG
 
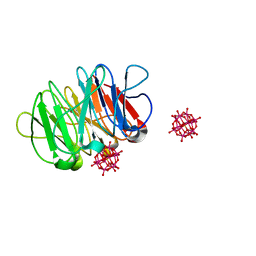 | | Crystal structure of the hybrid bioinorganic complex of Pizza6S and Keggin (STA) | | Descriptor: | Keggin (STA), Pizza6S | | Authors: | Noguchi, H, Vandebroek, L, Kamata, K, Tame, J.R.H, Van Meervelt, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hybrid assemblies of a symmetric designer protein and polyoxometalates with matching symmetry.
Chem.Commun.(Camb.), 56, 2020
|
|
1M6D
 
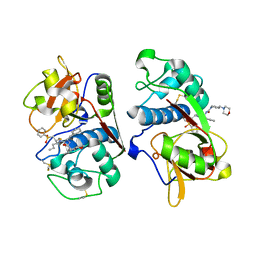 | | Crystal structure of human cathepsin F | | Descriptor: | 4-MORPHOLIN-4-YL-PIPERIDINE-1-CARBOXYLIC ACID [1-(3-BENZENESULFONYL-1-PROPYL-ALLYLCARBAMOYL)-2-PHENYLETHYL]-AMIDE, Cathepsin F | | Authors: | Somoza, J.R, Palmer, J.T, Ho, J.D. | | Deposit date: | 2002-07-15 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of human cathepsin F and its implications for the development of novel immunomodulators
J.Mol.Biol., 322, 2002
|
|
4OWC
 
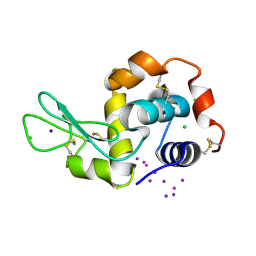 | | PtI6 binding to HEWL | | Descriptor: | CHLORIDE ION, IODIDE ION, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Starkey, V.L, Lamplough, L, Kaenket, S, Helliwell, J.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The binding of platinum hexahalides (Cl, Br and I) to hen egg-white lysozyme and the chemical transformation of the PtI6 octahedral complex to a PtI3 moiety bound to His15.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4OCW
 
 | | Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.06 heavy chain, CAP256-VRC26.06 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
1M4X
 
 | | PBCV-1 virus capsid, quasi-atomic model | | Descriptor: | PBCV-1 virus capsid | | Authors: | Nandhagopal, N, Simpson, A.A, Gurnon, J.R, Yan, X, Baker, T.S, Graves, M.V, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2002-07-05 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | The Structure and Evolution of the Major Capsid Protein of a Large,
Lipid containing, DNA virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M6E
 
 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
6QX3
 
 | | Influenza A virus (A/NT/60/1968) polymerase Hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
1N41
 
 | | Crystal Structure of Annexin V K27E Mutant | | Descriptor: | CALCIUM ION, SULFATE ION, annexin V | | Authors: | Mo, Y.D, Campos, B, Mealy, T.R, Commodore, L, Head, J.F, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interfacial basic cluster in annexin V couples phospholipid
binding and trimer formation on membrane surfaces
J.Biol.Chem., 278, 2003
|
|
4OC8
 
 | | DNA modification-dependent restriction endonuclease AspBHI | | Descriptor: | PHOSPHATE ION, restriction endonuclease AspBHI | | Authors: | Horton, J.R. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structure and mutagenesis of the DNA modification-dependent restriction endonuclease AspBHI.
Sci Rep, 4, 2014
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
4OD1
 
 | | Crystal structure of human Fab CAP256-VRC26.03, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.03 heavy chain, CAP256-VRC26.03 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
1MVT
 
 | | Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',4',5'-TRIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate Reductase, SULFATE ION | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W.A, Gangjee, A. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of two polymorphic forms of a pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolate binary complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N0T
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
1N44
 
 | | Crystal Structure of Annexin V R23E Mutant | | Descriptor: | Annexin V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Campos, B, Mealy, T.R, Commodore, L, Head, J.F, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interfacial basic cluster in anexin V couples phospholipid binding and trimer formation on membrane surfaces
J.Biol.Chem., 278, 2003
|
|
5M4X
 
 | |
1N4K
 
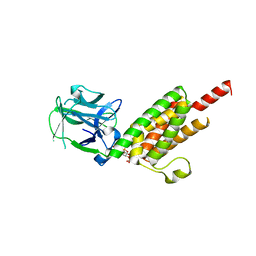 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Bosanac, I, Alattia, J.R, Mal, T.K, Chan, J, Talarico, S, Tong, F.K, Tong, K.I, Yoshikawa, F, Furuichi, T, Iwai, M, Michikawa, T, Mikoshiba, K, Ikura, M. | | Deposit date: | 2002-10-31 | | Release date: | 2002-12-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the inositol 1,4,5-trisphosphate receptor
binding core in complex with its ligand.
Nature, 420, 2002
|
|
6AVB
 
 | | CryoEM structure of Mical Oxidized Actin (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Grintsevich, E.E, Ge, P, Sawaya, M.R, Yesilyurt, H.G, Terman, J.R, Zhou, Z.H, Reisler, E. | | Deposit date: | 2017-09-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catastrophic disassembly of actin filaments via Mical-mediated oxidation.
Nat Commun, 8, 2017
|
|
5M0W
 
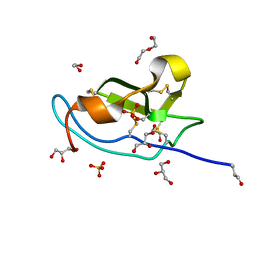 | | N-terminal domain of mouse Shisa 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lohkamp, B, Ojala, J.R.M. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ab initio solution of macromolecular crystal structures without direct methods.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LXW
 
 | |
6B77
 
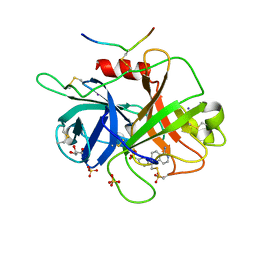 | | Structures of the two-chain human plasma factor XIIa co-crystallized with potent inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, GLYCEROL, ... | | Authors: | Dementiev, A.A, Silva, A, Yee, C, Flavin, M.T, Partridge, J.R. | | Deposit date: | 2017-10-03 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of human plasma beta-factor XIIa cocrystallized with potent inhibitors.
Blood Adv, 2, 2018
|
|
5TF2
 
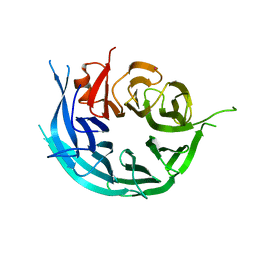 | | CRYSTAL STRUCTURE OF THE WD40 DOMAIN OF THE HUMAN PROLACTIN REGULATORY ELEMENT-BINDING PROTEIN | | Descriptor: | Prolactin regulatory element-binding protein, UNKNOWN ATOM OR ION | | Authors: | Walker, J.R, Zhang, Q, Dong, A, Wernimont, A, Li, Y, He, H, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Chen, Z, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-23 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF THE WD40 DOMAIN OF THE HUMAN PROLACTIN REGULATORY ELEMENT-BINDING PROTEIN (CASP target)
To be published
|
|
6B0R
 
 | |
4OWB
 
 | |
